Inherited Differences In Antibody Genes May Explain Variable Responses To Covid-19 Infection
(Posted on Friday, February 10, 2023)
Three years into the Covid-19 pandemic, it is clear that some of us produce stronger immune reactions to infection than others. Some may only experience mild flu-like symptoms, while others could be hospitalized or worse. Recent research suggests there is more to humans’ variable ability to control infection than pure chance. Common variations in immune-related genes heavily influence our immune reactions to invading pathogens, SARS-CoV-2 included.
Antibodies inherit their structure from the genetic Ig germline locus. The exact physical structure of the antibody is determined by the genetic predispositions of the patient, which is why one patient may develop much more effective antibodies than another, even if infected with the same virus at the same time.
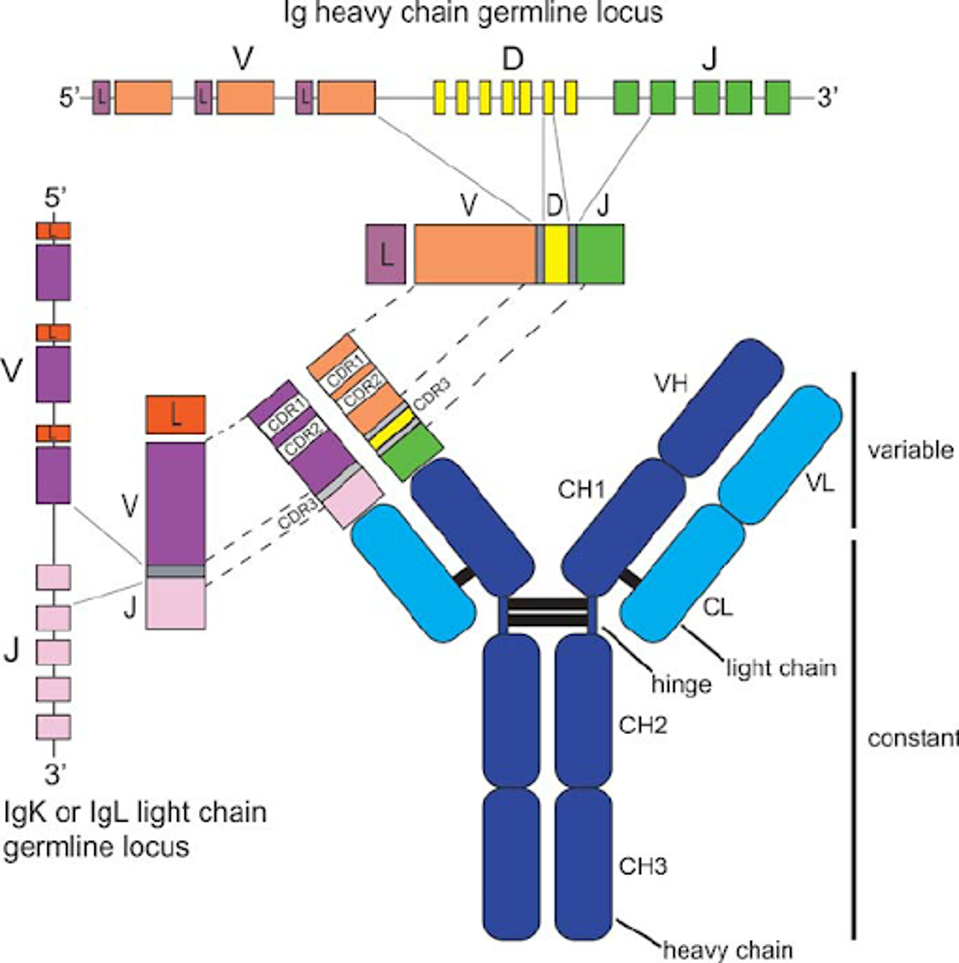
FIGURE 1: Antibody structure and genetic encoding.
SHILPA A JOSHI
Following exposure to Covid-19, our immune systems develop antibodies specifically tailored against the virus. These antibodies are highly variable in their efficacy. Pushparaj et al. shine some light on this variation. They find that minor differences in our genes and in the antibody genes that we inherit may also influence the structure and efficiency of the antibodies we develop post infections. Here we discuss the implications of their findings and how we may use this data to inform antibody discovery efforts better moving forward.
Measured Variation in Response to Covid-19 Infection
The researchers extracted the sera of 14 healthcare workers seven months after their infections in May 2020. Every patient had developed antibodies towards SARS-CoV-2 but in a wide array of genetic diversity. Antibodies are encoded by B-cell receptor sequences, which often vary significantly, leading to the significant genetic diversity of produced antibodies.
From the sera, they analyzed individual antibodies. From these antibody structures, they could determine the exact inherited germline, meaning every individual genetic detail of the sequence that would encode the antibody. By analyzing each allele individually, they could compare for relative presence in neutralizing antibodies.
They found that the number of encoded alleles tallied between 44 and 61 per patient. One of the most critical alleles to antibody potency, IGHV1-69, was found to have six variants among the 14 patients: IGHV1-69∗01, ∗02, ∗04, ∗06, ∗09, and ∗20. This will be the focus of Pushparaj et al.’s study moving forward, as they aim to understand the impact of gene polymorphisms on antibody efficacy.
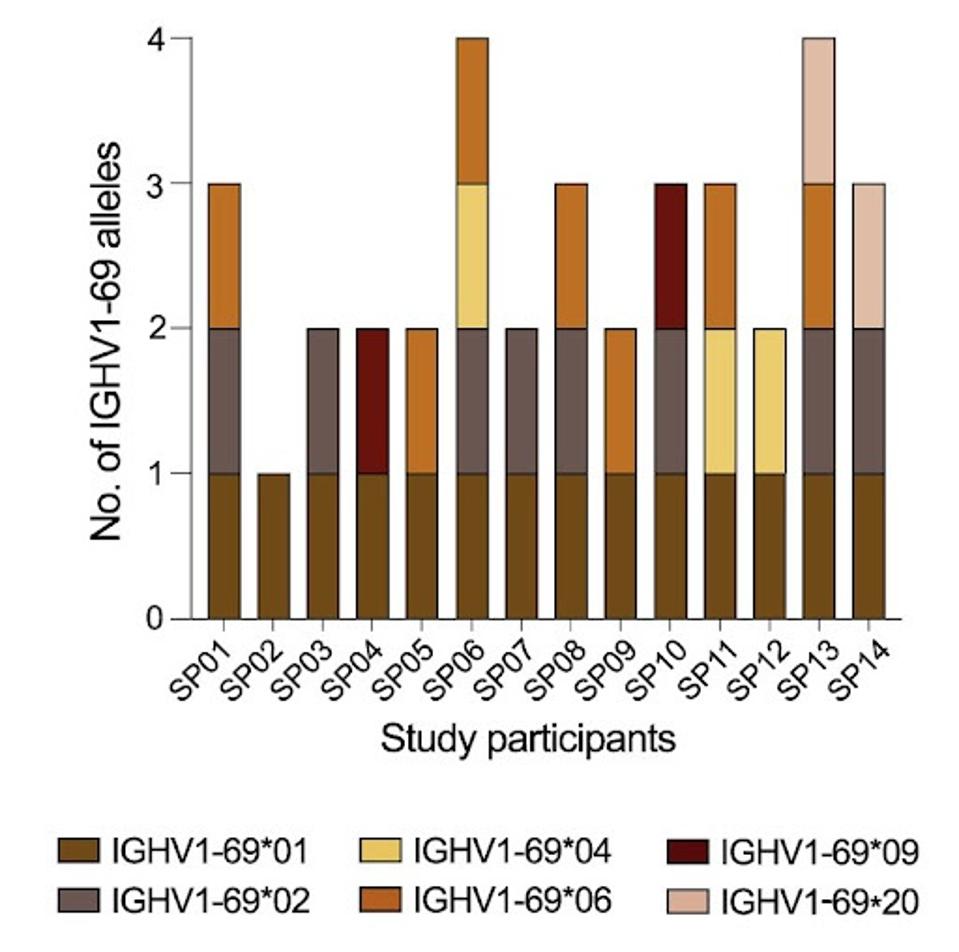
FIGURE 2: A summary plot showing which IGHV1-69 alleles were present in each study participant.
PUSHPARAJ ET AL.
Antibodies Efficacy is Related to Genetic Variation
Using the sera of SP14, a patient who displayed the IGHV1-69∗01, IGHV1-69∗02, and IGHV1-69∗20 alleles, the researchers generated several heavy and light chains. These are the building blocks of antibodies. From this sole patient, they generated 29 spike-specific monoclonal antibodies, 15 of which were neutralizing against the wildtype SARS-CoV-2. All those that neutralized bound the receptor-binding domain of the virus.
Notably, most neutralizing antibodies used a variation of IGHV1-69, specifically IGHV1-69∗02 or IGHV1-69∗20. To reiterate, these patients had alleles between 44 and 61 each. Still, a variation of IGHV1-69 was found in a large majority of neuralizing antibodies in this particular patient, reinforcing the notion that this specific allele is critical to developing potently neutralizing antibodies in the immune system.
However, IGHV1-69 has allelic variation. Each variant allele may contain one or more somatic hypermutations that characterize it differently from others. These minor genetic variations in the alleles once encoded onto a monoclonal antibody, may account for some or all of the neutralizing capacity of the antibody, or lack thereof.
IGHV1-69 Allele Usage Influences Neutralizing Antibody Activity
They next tested the value of somatic hypermutations (SHMs) in the allele to the final neutralization of the produced antibody. In other terms, do mutations in the important alleles make a difference? Pushparaj et al. found that by removing SHMs from the IGHV1-69∗20 allele found in patient SP14, the resultant antibody lost neutralizing capacity against SARS-CoV-2.
They repeated this process with all other variations of IGHV1-69 and found similar results. In essence, the SHMs are critical to neutralizing capability.
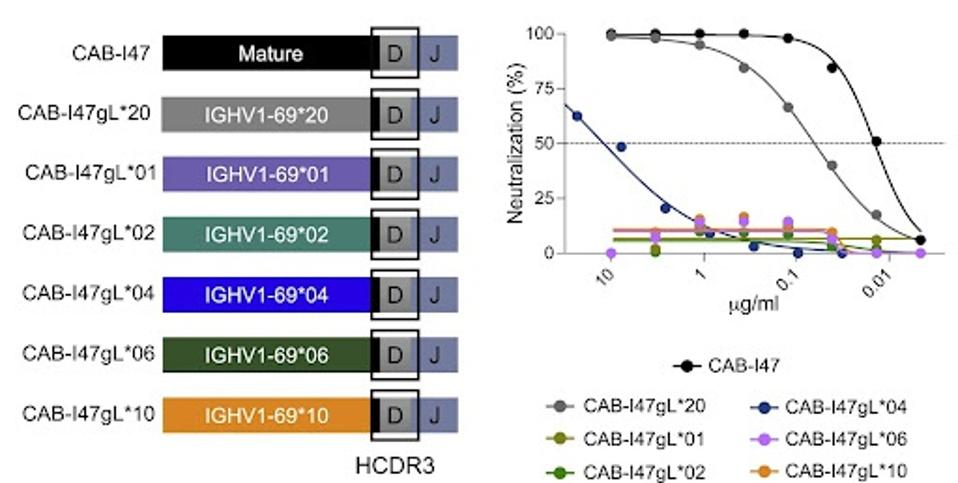
FIGURE 3: Neutralizing activity of the variant mAbs against the ancestral SARS-CoV-2 strain is shown as curves (middle) and IC50 values (μg/mL).
PUSHPARAJ ET AL.
Structural Analysis Reveals the Basis for IGHV Allele Requirement
To understand further why certain somatic hypermutational differences in specific alleles are so important, the researchers conducted a cryo-electron microscopy analysis of the previous antibody in question: CAB-I47. They wanted up close the binding differences between the antibody produced from the SHM and non-SHM alleles.
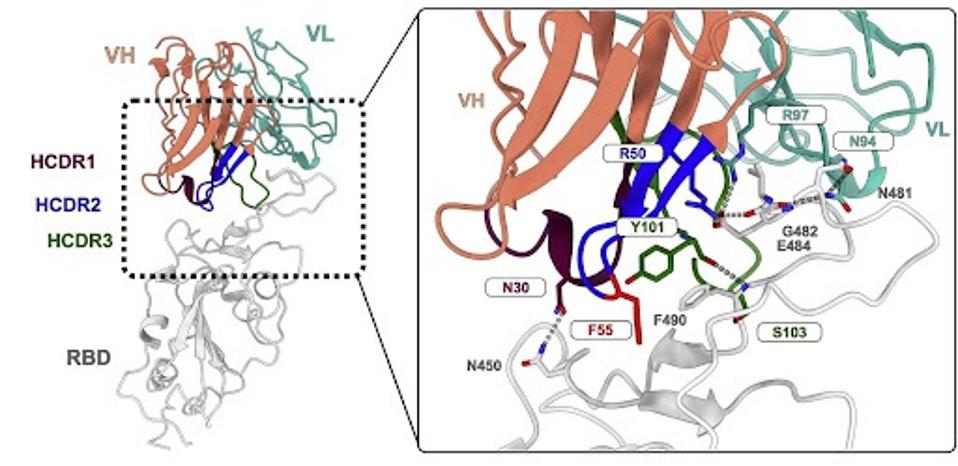
FIGURE 4: Molecular details of the CAB-I47 Fab and RBD interaction with residues important for the interaction labeled.
PUSHPARAJ ET AL.
As with many monoclonal antibodies, CAB-I47 makes contact with the receptor-binding domain to block binding to the human ACE2 receptor. There are a few critical contacts between the antibody and the receptor-binding domain, most notably the interaction between R50 of the antibody and E484 and G482 of the receptor-binding domain.
One amino acid can make a big difference. A single amino acid variation in the inherited structure may yield a significant difference in antibody binding and neutralization. R50 is variable among the different versions of IGHV1-69. Some mutate the arginine (R) to glycine (G), while others leave it unmutated. The R50G mutation seems to be the culprit for the loss of neutralizing activity, as the unmutated version forms hydrogen bonds and salt bridges between the virus and antibody.
Discussion
This is but a single example of one minor variation in the antibody germline completely reducing the neutralizing capacity of an antibody candidate. There are likely thousands, if not millions, of similarly crucial allele modifications that can make or break a potential monoclonal antibody. Not only can minor variations negatively impact an antibody, but it would follow that such minor variations could also drastically improve neutralizing efficacy.
How can we identify and maximize these minor differences to our advantage? We recently detailed an artificial intelligence mechanism designed to do just that; it identifies and tests minor antibody variations to identify the most potent germline sequence possible. It is conceivable that genetic allele variation could be incorporated into that mechanism to optimize the system further. We can only hope such a process will be implemented shortly as hundreds of people succumb to Covid-19, desperately needing effective monoclonal drugs to fight this disease.

