New Tanzanian Variant Detected In Angola From An Entirely New Branch Of SARS-CoV-2
(Posted on Thursday, April 15, 2021)

Giraffe in front of Kilimanjaro
WWW.IEXPLORE.COM
The versatility of SARS-CoV-2 to evolve new variants that increase transmissibility, virulence, and immune evasion is a new troubling feature of the Covid-19 pandemic. The recent discovery of a novel variant emerging from Tanzania adds a new chapter to this disturbing story. Up until the discovery of the new variant, all other variants of interest or concern derive from a common ancestral virus, the B.1 strain that first made its appearance in early 2020. This is not so for the newly described variant. It evolved from an entirely different source, the A lineage, a finding that substantially expands our understanding of the repertoire of mutants we must be prepared to contend with in the months and years ahead.
The difference between the A and B lineages are three mutations that have come to define the B lineage that has displaced almost all others around the world. The common understanding is that a single amino acid change in the spike protein, the D614G mutation, increases both the ability of the virus to bind to the ACE2 receptor and, at the same time, stabilizes the interaction between the S1 and S2 protein of the spike, conferring an increase in transmissibility. I and others have suggested that the D614G substitution may not be all to the story of success for the B.1 variant. The B.1 linage viruses carry two additional mutations: the P323L mutation in the RNA-dependent RNA polymerase (NSP12), which is the key to virus replication and the production of viral mRNA and another mutation in the 5 prime untranslated region of the genome. Although hardly studied, both of these mutations may contribute, along with D614G, to the replication competence and transmissibility in the spike protein.
The Tanzanian variant (which is how I will denote it as it lacks official designation) teaches us that variants of interest and concern may lack all three defining mutations of the B.1 strain. Nonetheless, the new variant is of interest and of possible concern as it carries a number of mutations in the spike protein characteristic of other bonafide variants of concern from the B.1 lineage. Of the 13 mutations that distinguish the spike protein of the Tanzanian variant from the original Wuhan strain, eight are found in the B.1 family of variants. This is a remarkable illustration of convergent evolution. No one B.1 variant carries all these mutations, but each must confer some selective advantage to the A lineage variant. It is worth noting the five spike mutations unique to the Tanzanian virus, as they are likely to appear sooner or later in B.1 linage variants as well.
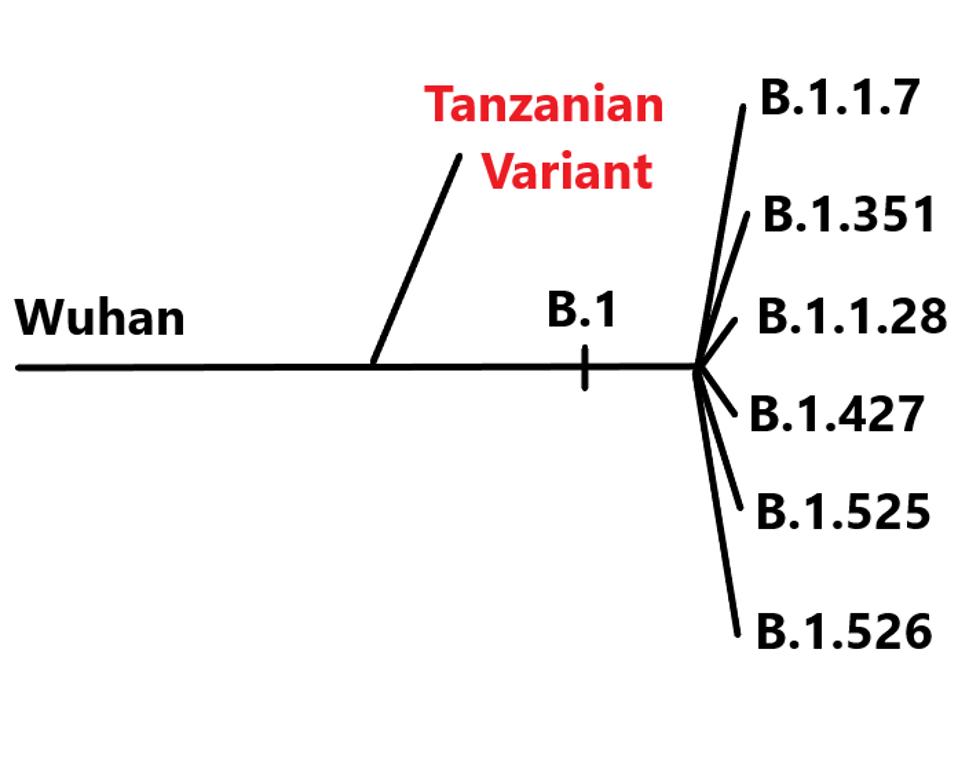
Tanzanian Variant lineage is reference to other variants
The observation of greatest concern is that the Tanzanian virus posses the E484K mutation found in many of the B.1 variants of concern. This mutation confers resistance to neutralizing antibodies of convalescent and vaccine plasma and also reduces the activity of some neutralizing monoclonal antibodies. This observation suggests that like other variants, this Tanzanian virus may heed resistant, at least partially, to the current generation of Covid-19 vaccines. The E484K mutation also increases the affinity of the spike protein for the ACE2 receptor, raising the possibility of increased transmissibility.
Another mutation of note in the spike protein is P681H. This change occurs near the cleavage site between the S1 and S2 spike subproteins. We and others speculate that this mutation increases transmissibility by increasing the efficiency of the spike precursor S protein, a requirement for infectivity.
All of the variants of concern carry a number of mutants in the N-terminal domain of the spike protein. Again, in an example of convergent evolution, four of these mutations are present in B.1 lineage variants, but not all together in any other single variant. The N-terminal domain contains a super antigenic site, the target of many neutralizing antibodies. Many of the N-terminal domain mutants of the Tanzanian virus map to this site.
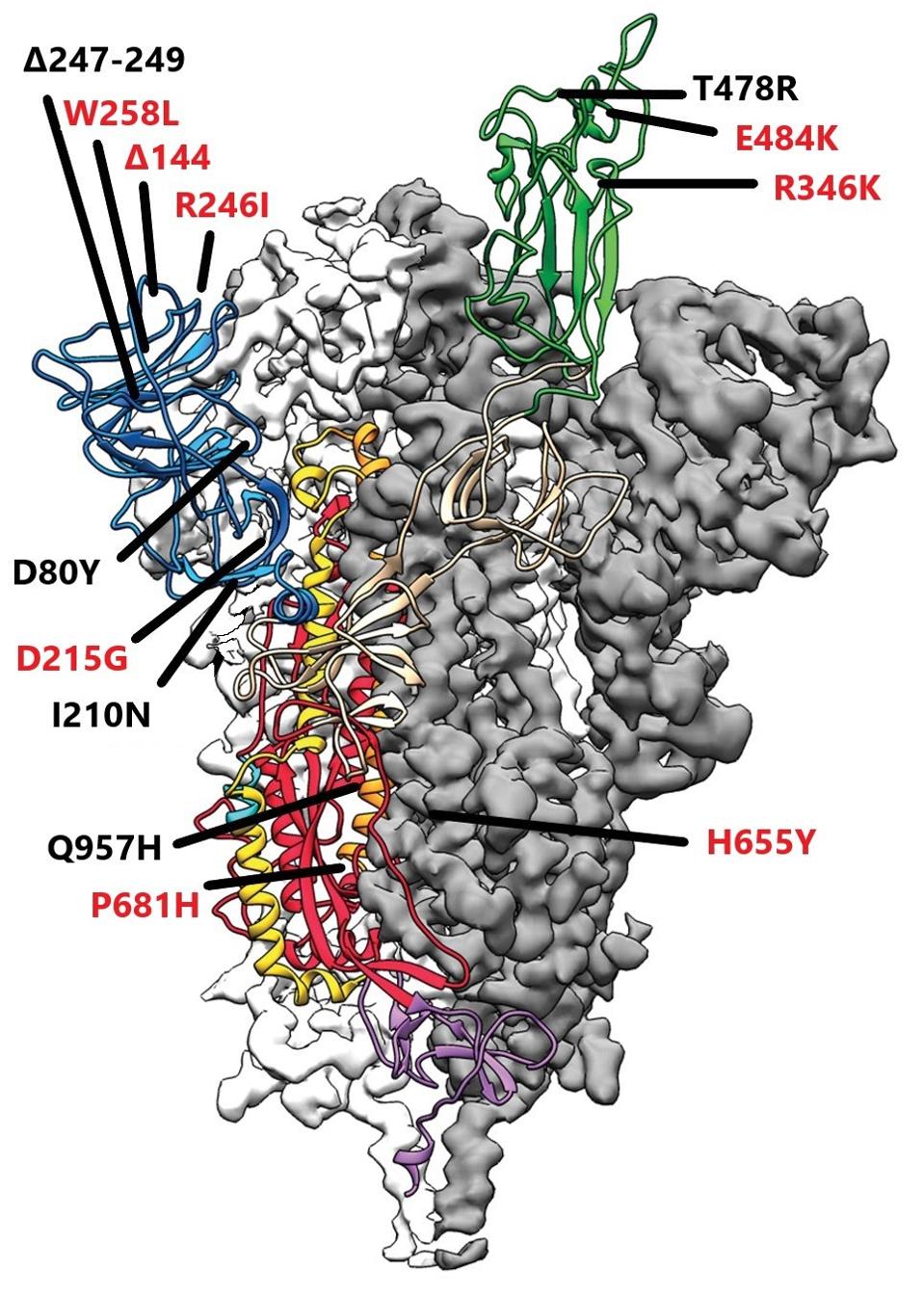
Tanzanian Variant spike protein: Mutations in red are those found in “B” lineage variants; mutations in black are those that are unique to the Tanzanian variant
AMERICAN CHEMICAL SOCIETY
As no specific function is attributed as yet to the N-terminal domain, this set of mutations is thought to increase the ability of the virus to escape immune detection. I suspect that is not all there is to the story. We can ask the question: Why are there such convergence-specific mutations in the superantigenic and other sites? Investigations of the functions of the N-terminal domain deserve further study.
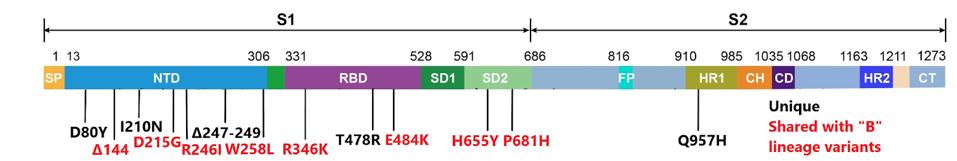
FIGURE 2 SPIKE MUTATIONS: Δ144 is shared with B.1.1.7 and B.1.525; D215G is shared with B.1.351 and B.1.525; R246I is shared with B.1.351; W258L is shared with B.1.427/9; R346K is shared with escape variants in India (no designation); E484K is shared
An additional 18 amino acid changes occur in proteins outside the spike protein. These include 14 in the orf1ab proteins that specify the replication complex. Extensive mutations in the replication complex are a common feature of B.1 linage variants. I and others speculate that these mutations confer enhanced replication competence, contributing to an increase in viral load virulence and transmission. Not all of the selective advantage of variants is readily attributed to mutations in the spike proteins alone.
Of the remaining four amino acid changes outside of the spike protein, there are two in the structural proteins, one each in the envelope (E) protein and nucleocapsid (N) protein. Antibodies targeting both are present in convalescent plasma raising the possibility that these changes contribute to immune evasion. The remaining two mutants both change the coding sequence of the accessory orf8 protein. One of these is identical to a mutation in a B.1 linage variant of concern. I note that orf8 is amongst the most frequently mutated viral proteins common to all variants of concern. The orf8 protein is one of the most antigenic of the SARS-CoV-2 proteins, raising the reasonable hypothesis that these are escape mutants. Elsewhere, some have suggested that orf8 may slow viral replication and that mutations that truncate the orf8 protein otherwise reduce its function may increase the viral load in infected patients.
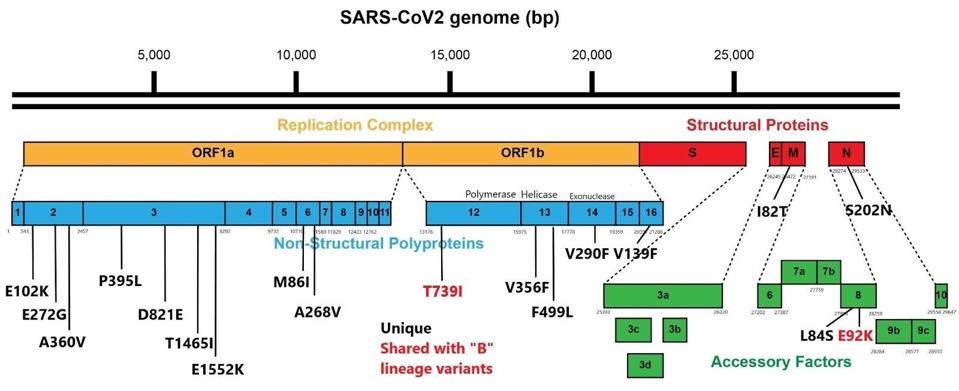
FIGURE 3 EXTRA-SPIKE MUTATIONS: T739I in NSP12 is shared with B.1.1.298; E92K in orf8 is shared with B.1.1.28.1; all other mutations external to the spike protein are unique
So why is this new variant relevant? This is another variant we have to be especially vigilant towards. It may be as infectious and immune evasive as the widespread B.1.1.7 and B.1.351 variants, in addition to potential reinfection capabilities, as they carry similar mutations. It also represents why we need more comprehensive variant surveillance around the world. Were this not detected in the Angolan airport, it may have caused havoc in the country and elsewhere.
We do not yet know exactly how dangerous the Tanzanian variant may be. That is partly due to the new total blackout of information during the pandemic from that country. Official data would have that no infections have occurred in Tanzania since May 2020, which is clearly not the case. The appearance of a new variant of interest and possibly of concern detected in travelers from that country highlights the need for transparency, both for the control of Covid-19 within the country and for the dangers viruses emanating from Tanzania may pose for the rest of the world.
Read the full article on Forbes.
Originally published on April 15, 2021.

