An Antibody Cocktail To Lay Low A Mighty Foe
(Posted on Thursday, April 29, 2021)
Monoclonal antibodies have proved effective in the prevention and treatment of Covid-19. Their effectiveness depends on the recognition of specific structures on the surface of the viral spike protein. Over the past six months, we have learned that many of these shape-specific determinants change in ways that abrogate the effectiveness of individual antibodies and even of antibody cocktails. This is the second in a series that describes a search for monoclonal antibodies that may successfully address the problem of antigenic variation. Read more from this series in part one.

Statue of Achilles with an arrow through his heel.
FILIPPO ALBACINI, “THE WOUNDED ACHILLES”
Viruses like SARS-CoV-2, influenza, and respiratory syncytial virus, are escape artists. These viruses evade immune recognition by mutations that reduce immune recognition yet preserve the ability to infect and cause disease.
SARS-CoV-2 changes many sites of the exterior spike protein to avoid neutralization, all while maintaining replication competency and in some cases, boosting competency and receptor binding avidity. However, there are some amino acids of the spike protein that are not often changed. These conserved areas may be an Achilles’ heel of the virus. Understanding these conserved regions is vital to both therapeutic and prophylactic antibody design, as well as design for future generation coronavirus vaccines.
In a recent paper, Starr et al. describe SARS-CoV-2 antibodies that neutralize not only the B.1 virus but also all variants tested as well as SARS-CoV, several bat coronaviruses and even a bat coronavirus that binds to a different receptor.
Most therapeutic antibodies in use today were isolated based on avidity for the SARS-CoV2 receptor, the cell surface protein ACE2, and for neutralization potency. In contrast, the Starr et al. antibodies were selected for both neutralization and their ability to neutralize a diverse set of coronaviruses, most of which bind to the ACE2 receptor. The breadth of neutralization rather than potency was a primary selection criterion.
In a series of experiments, they determined the binding sites of each of twelve antibodies on the SARS-CoV-2 receptor-binding domain (Figure 4). They also developed a detailed map of possible escape variants of each amino of the entire domain (Figure 3). Finally, they measured the ability of each antibody to neutralize a broad set of SARS-CoV-2 variants of concern as well as an ACE2 binding, including SARS-CoV and one non-ACE2 binding coronaviruses of diverse origin (Figure 2).
The twelve antibodies studied naturally sort themselves into three classes, those that bind to the region of the receptor-binding domain that interacts directly with the ACE2 receptor, those that bind near the ACE2 binding (the receptor-binding motif), and a third category that binds to a distal region named the core region.
Antibodies targeting the receptor-binding motif and ACE2 contact epitopes neutralize SARS-CoV-2 effectively but fail to effectively neutralize many of the variants and all of the non-SARS-CoV-2 viruses. There is one exception, S2E12, which both neutralizes a broad range of viruses and which is unaffected by almost all amino acid substitutions in the receptor-binding domain. The S2E12 exception is attributed to the recognition of contacts between the subunits of the trimeric receptor that are constrained to maintain consistent protein interactions even though individual amino acids at the contact points may vary.
The core receptor-binding domain antibodies bind to sites distant from ACE2 binding have lower potency than those which bind to the receptor-binding motif. However, these antibodies typically neutralize most variants and non-SARS-CoV-2 viruses (Figure 2). The binding and neutralization of these antibodies are impervious to most amino acid substitutions in the receptor-binding domain.
As an analogy, think of the receptor-binding domain having a mushroom shape, with a cap and a stem. In this analogy, the ACE2 binding site occupies the top of the cap. Some of the conserved sites occur on the stem and to some extent the edges of the cap. Specifically, S309 and S2X35 bind high up on the stem and the underside of the cap. Others including S304 and S2H97, bind on the lower parts of the stem (Figure 1).
The four most effective core receptor-binding domain antibodies were S2H97, S304, S309, and S2X35. These binding sites of the antibodies are pictured in Figure 1. The part of the ACE2 receptor to which the spike binds is colored pink. The intensity of the color blue of the receptor-binding domain denotes how well the region tolerates amino acid substitutions. Notice that the base of the receptor domain is highly conserved.
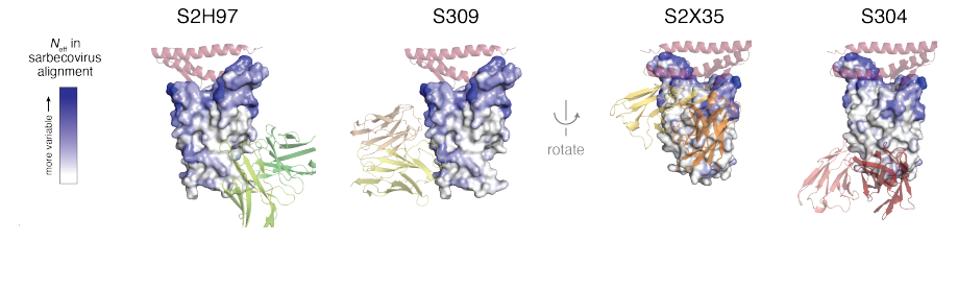
FIGURE 1: Four receptor-binding domain core antibodies
STARR ET AL.
The four antibodies shown in figure 1 neutralize SARS-CoV-2, most of the variants of concern tested, as wells many non-SARS-CoV-2 coronaviruses. All, but S309, effectively neutralize the Kenyan bat coronavirus BtKY72. S2H97 and S304 effectively neutralize European bat coronavirus BM48-31. S2H97 takes it one step further than the rest and also neutralizes a number of Asian coronaviruses that use a different receptor altogether (Figure 2).
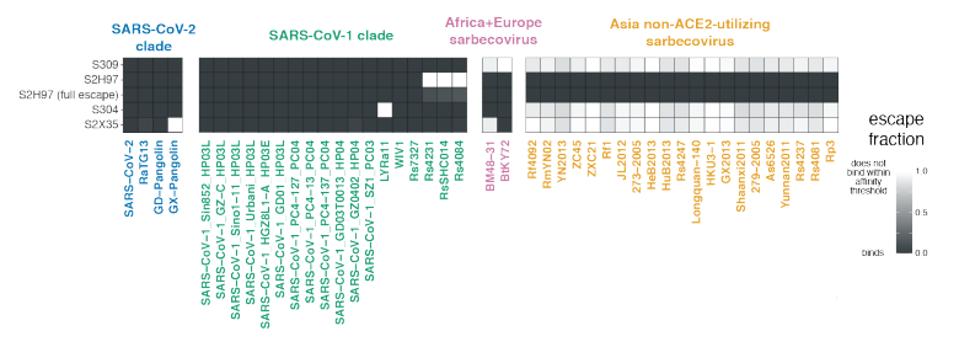
FIGURE 2: Four core antibodies binding to non-SARS-CoV-2 viruses.
STARR ET AL.To generate the data in figure 3, amino acids in the spike protein receptor-binding domain were changed to every other possible amino acid. The majority which remain functioning were then tested to examine whether or not the spike, with these mutations, could be neutralized. In every case, some laboratory-generated mutations are capable of eliminating antibody binding. The letters denote the amino acid substitutions at that site and the height represents the reduction in the binding capability of the antibody. For example, S2H97 has reduced binding when E516 and L518 mutations are introduced, whereas those mutations can be sustained by S309.
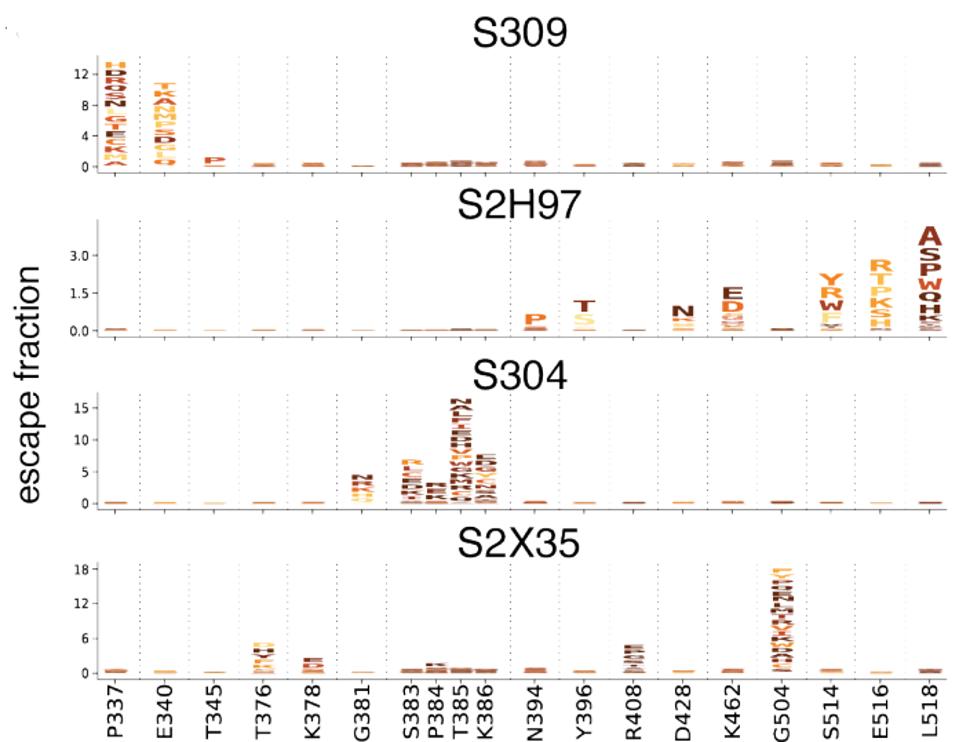
FIGURE 3: Escape mutants from the four core antibodies.
STARR ET AL.Each antibody recognizes epitopes that, if mutated, render the virus resistant to neutralization. For S309, mutations to P337 and E340 confer 12-fold increase in resistance. For S2H97, mutations to S514, E516, and L518 can reduce neutralization three-fold. For S304, mutations to S383, T385, and K386 result in 15-fold or more reduction. Finally, for S2X35, mutations to G504 can induce neutralization 18-fold or more. resistance. While these mutations have not appeared in variants of concern and are relatively rare due to the region in natural isolates, they do still appear in GISAID database sequences, ranging between 9 and 759 viruses (Table 1).
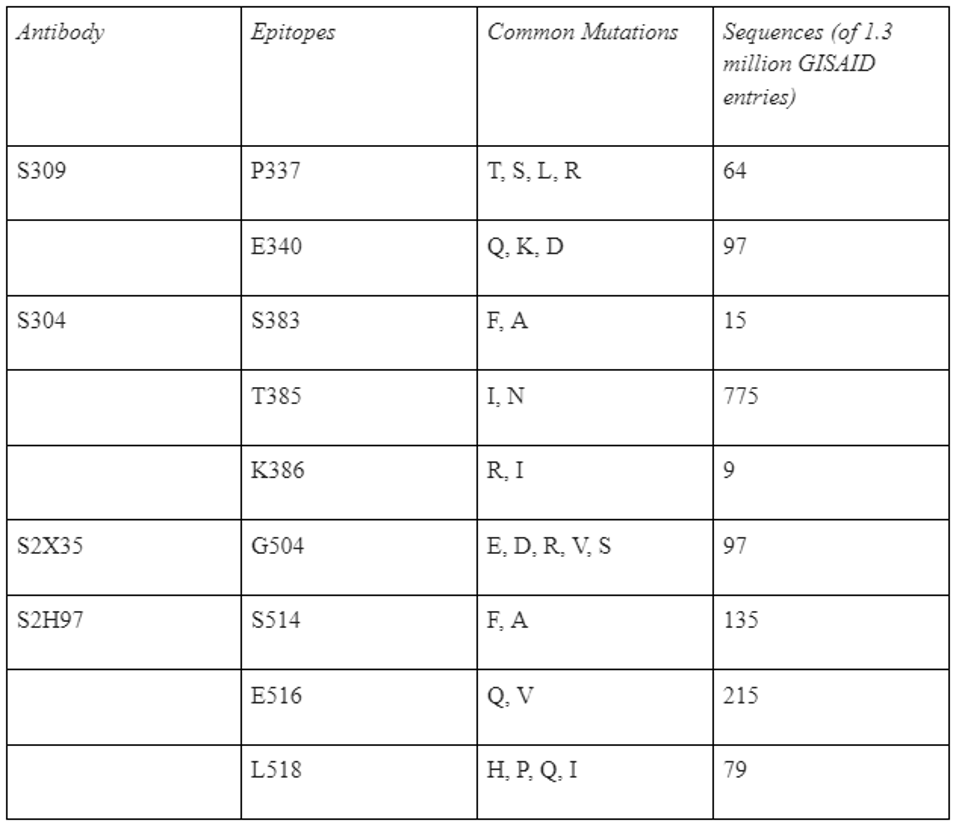
TABLE 1: Table of escape mutations for core antibodies.
HASELTINEFigure 4 depicts the binding footprints of the antibodies on the receptor-binding domain of SARS-CoV-2 of all twelve of the antibodies analyzed in this study. The black areas are those regions in which it appears that amino acid changes result in substantial loss of receptor binding domain stability and/or function. The red areas are regions of the receptor-binding domain where amino acid changes are functionally tolerated, but substantially reduce the binding of the antibody to that region.
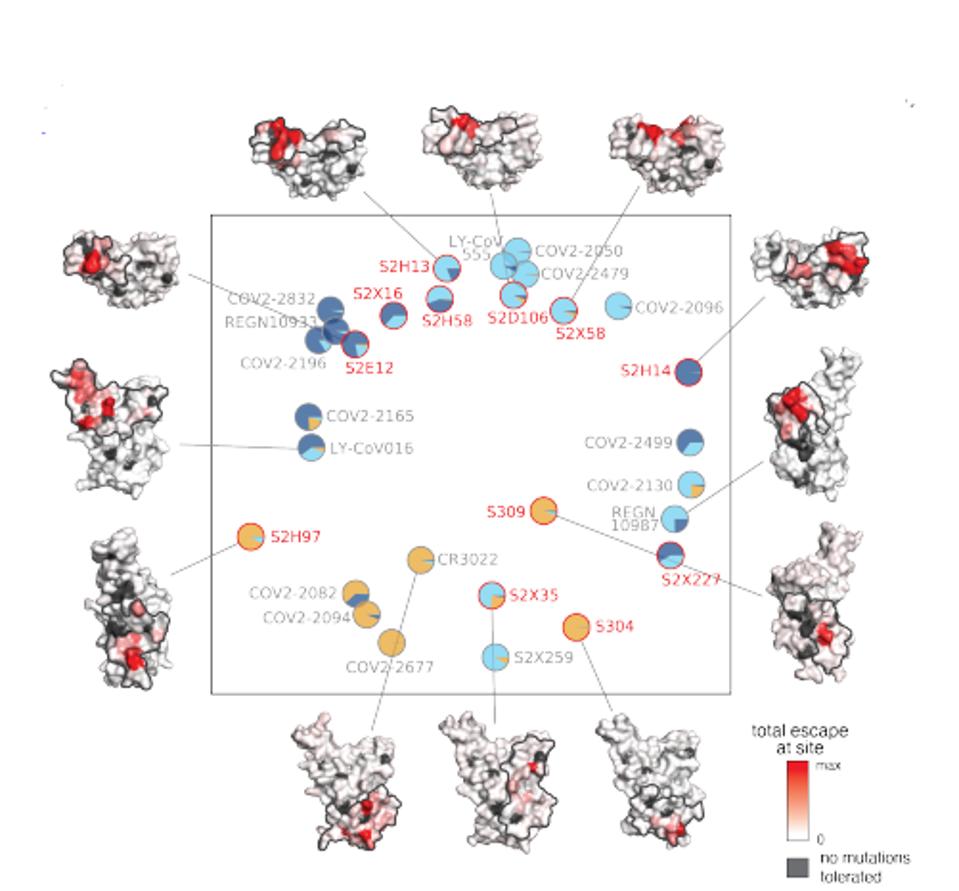
FIGURE 4: Twelve antibodies in this study.
STARR ET AL.
This research provides a blueprint for future anti-SARS-CoV-2 antibody drug development. Some of these antibodies are impressive on their own, such as S309, the basis for VIR-7831 and VIR-7832 drug candidates currently in clinical development. Cocktails containing two or more of these antibodies may be even more effective in countering resistance and neutralization of variants than will any one alone. I suggest an ideal cocktail would contain two bi-specific antibodies covering all four conserved core footprints. It might help to add the modifications in the Fc portion of these antibodies that increase the half-life of the antibodies, mucous membrane transport, and antibody-mediated cytotoxicity as was done to create the in the VIR-7831 and VIR-7832 drug candidates.
Conservation of the “mushroom stem” binding sites amongst distantly relates coronaviruses does necessarily not mean they are immutable. There are two potential interpretations of such conservation. The first is that there is no selective pressure to change because the virus has been successful in diverting immune attention away from these conserved regions, specifically to the ACE2 binding site and to epitopes of the N-terminal domain of the S1 protein. The second is that the conserved areas are functionally relevant and cannot be changed. The first interpretation may have merit, as one report noted that the cold-causing coronavirus’s receptor-binding domain naturally varies in amino acid composition by as much as 17% and yet remains functional.
An additional caution: The Starr et al. studies are restricted to the receptor-binding domain, a small but critically important region of the entire S protein. The development of antibodies to the receptor receptor-binding domain alone may not be the only winning strategy. Mutations in the spike protein within and outside of the receptor-binding domain as well as in the extra-spike proteins of the virus may have compensatory effects. Decreases in viral infectivity and transmission lost by changes in the conserved regions may be made up for by increases in the activity of the replicative complex, the stability of the virus particle, and increases in the activity of the accessory proteins. Indeed, all variants characterized to date have more mutations in proteins other than the spike than they do in the spike protein itself. Infectivity, pathogenesis, and neutralization are composite properties of all viral functions.
These and other studies may have found at least one Achilles’ heel of SARS-CoV-2. I doubt that will be the end of the story for this wily escape artist. In time believe we will finally throttle this many-headed hydra with a combination of antiviral drugs, each targeted to a different essential viral protein, combined with a cocktail of antibodies such as those so elegantly described by Starr et al.
Read the full article on Forbes.
Originally published April 29, 2021.

