A New Twist To Antibody Cocktails To Prevent And Treat Covid-19
(Posted on Thursday, May 13, 2021)
Monoclonal antibodies have proved effective in the prevention and treatment of Covid-19. Their effectiveness depends on the recognition of specific structures on the surface of the viral spike protein. Over the past six months, we have learned that many of these shape-specific determinants change in ways that abrogate the effectiveness of individual antibodies and even of antibody cocktails. This is the fifth in a series that describes a search for monoclonal antibodies that may successfully address the problem of antigenic variation. Read more from this series in parts one, two, three, and four.

Monoclonal antibodies have been successful in both prevention and early treatment of Covid-19. The monoclonal antibodies that have been studied to date bind to and interact with the spike protein, and more specifically, the receptor-binding domain. Over the past several months, as reviewed in this series, new monoclonal antibodies have been discovered that have broad neutralizing capabilities. All of these antibodies bind to the receptor-binding domain, either to the receptor-binding motif, which interacts directly with the ACE2 receptor, or to other parts of the receptor-binding domain to inhibit function.
Despite the broad activity of these antibodies or camelid nanobodies, none are entirely neutralizing. SARS-CoV-2 is capable of mutating, leading to viral variants which inhibit antibody function that escape neutralization, triggering a search for effective neutralizing antibodies that recognize epitopes in regions outside the receptor-binding domain which might be constrained functionally, and therefore, less likely to mutate.
In a new study by Garrett et al., a complete set of overlapping linear peptides on a phage display backbone was developed, which spanned the entire spike protein. For reference, the receptor-binding domain is only about 15% of the full spike protein, stretching from amino acid 332 to 523 (Figure 1).
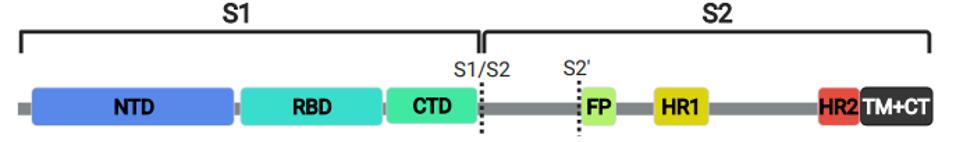
FIGURE 1: SARS-CoV-2 Spike Protein. N-terminal domain (NTD); receptor-binding domain (RBD); carboxy-terminal domain (CTD); fusion peptid (FP); heptad repeat (HP); transmembrane and carboxy terminus (TM+CT)
GARRETT ET AL.To examine naturally occurring antibodies that recognize these epitopes, they identified those linear regions which are recognized by convalescent sera. The figure below shows the regions in which naturally occurring convalescent antibodies bind linear epitopes (Figure 2). It is important to note that these are very different from what is seen when the entire spike protein or receptor-binding domain is used as a target. When the spike protein or receptor-binding domain are targeted, the recognized structures are three-dimensional and yield different binding actions. Whereas in this study, linear epitopes are recognized and the receptor-binding domain is nearly ignored. This is notable as the receptor-binding domain is the principal source for neutralizing monoclonal antibodies in most previous studies. Additionally, antibody responses vary from patient to patient, as no two immune responses are identical.
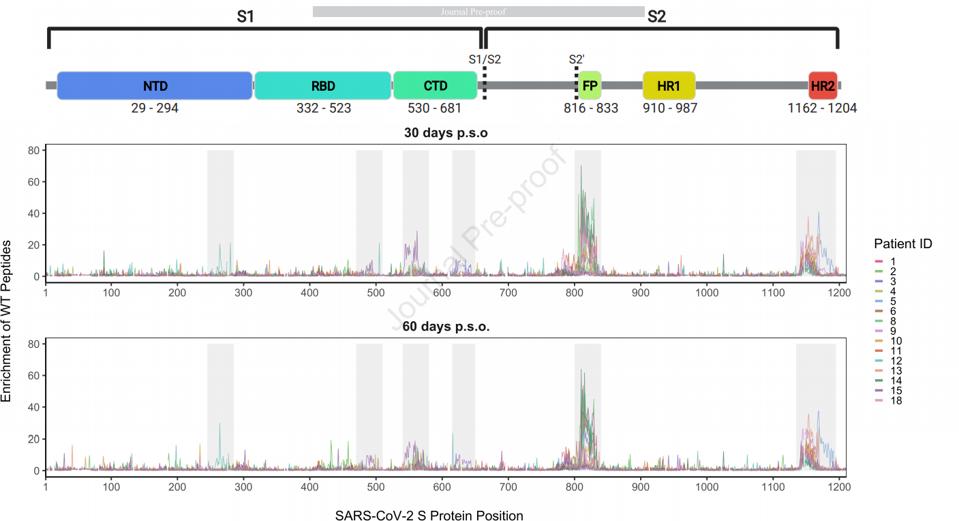
FIGURE 2: Major regions of antibody binding
GARRETT ET AL.The antibodies were next exposed to the linear epitopes and examined their ability to neutralize a pseudotype virus. Their samples indicated that 0% to 59% of the neutralization activity present in patient plasma is directed at non-receptor-binding domain epitopes. After depleting the neutralizing antibodies targeting the receptor-binding domain, the researchers found that non-receptor-binding domain antibodies, while not completely responsible for virus neutralization in patients, are at least partially responsible for residual neutralization, and are therefore important to the neutralization process.
The linear non-receptor-binding domain epitopes of interest map into four specific regions. These are the carboxy-terminal, the n-terminal domain, the fusion peptide, and the carboxy-terminus of the S2 subunit. To grasp whether these antibodies could tolerate amino acid mutations in these regions, the researchers developed a heat map of every mutation to individual positions. For example, in the figure below for the fusion peptide, the red indicates a reduction in binding affinity, and the blue indicates a boost. The intensity of the color shows the magnitude of reductions or promotions (Figure 3).
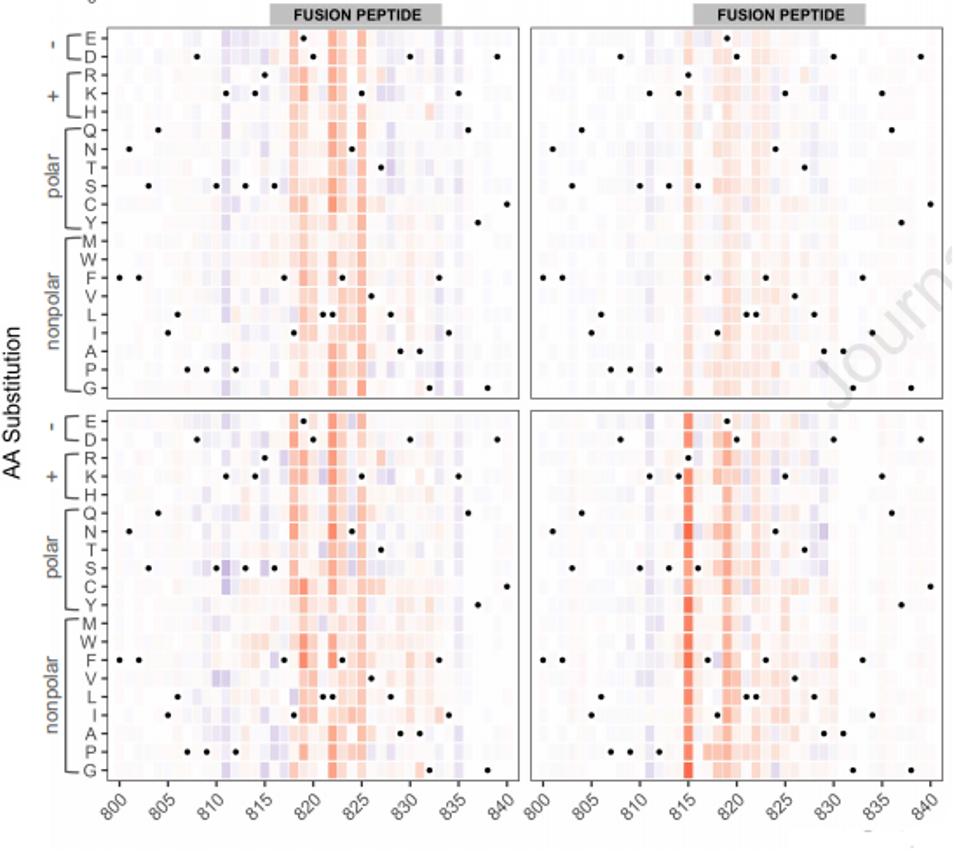
FIGURE 3: Heat map of positional mutations affecting antibody binding
GARRETT ET AL.Using this technique, they were able to divide these regions into two classes: those in which amino acid mutations in the region affect binding and those that are relatively resistant to amino acid mutations. It is this second class that provides the most interest. These results may have identified linear regions outside the receptor-binding domain that are constant regardless of variation, most likely because they are required for a conserved function.
In summary, these results suggest that it would be valuable to add to the current cocktails that recognize the receptor-binding domain. One or two additional antibodies that recognize highly conserved epitopes, perhaps in the vicinity of the fusion peptide, may be most useful, as this region was conserved across many variants and viruses. Additionally, we have commented that highly effective cocktails for treatment and prevention might be combined with combination chemoprophylaxis with alternate drugs for maximum effect, creating multiple targets on the spike protein for neutralization (Figure 4). It is unlikely that variants can quickly arise that are resistant to antibody and drug cocktails simultaneously. As new variants spread through populations, this combination strategy may be successful in the prevention of new variant-associated outbreaks.
Read the full article on Forbes.
Originally published on May 13, 2021.

