How SARS-CoV-2 Evades And Suppresses The Immune System (Part 3)
(Posted on Monday, August 16, 2021)
This is the third article in a 15-part series called “Relevance of Immune Suppression by SARS-CoV-2 to Understanding and Controlling the Covid-19 Pandemic,” which will explore an underappreciated but highly significant aspect of SARS-CoV-2 replication. The ability of SARS-CoV-2 to delay, evade, and suppress the immune system has myriad implications for drugs, vaccines, and other aspects of our pandemic response. The first set of pieces in this series are intended for a general audience; the second set, for the medical community; and the third and final set, for biomedical researchers looking for a deeper understanding of variants, how they’re generated, and what we might do to control them. Read part one and part two.

A classic example of a Rube Goldberg machine. Note how many pieces and processes need to come together for the light at the end to illuminate. Block one and the light won’t turn on.
GETTY
A complex genome and replication strategy offers many new opportunities to develop drugs that prevent and treat SARS-CoV-2 infections.
The SARS-CoV-2 genome is one of the largest of any RNA virus. On either side of the sequence are 5 prime and 3 prime untranslated regions, necessary for making new copies of the virus genome and initiating messenger RNA synthesis. The untranslated regions also contribute to control of viral replication and messenger RNA synthesis and translation.
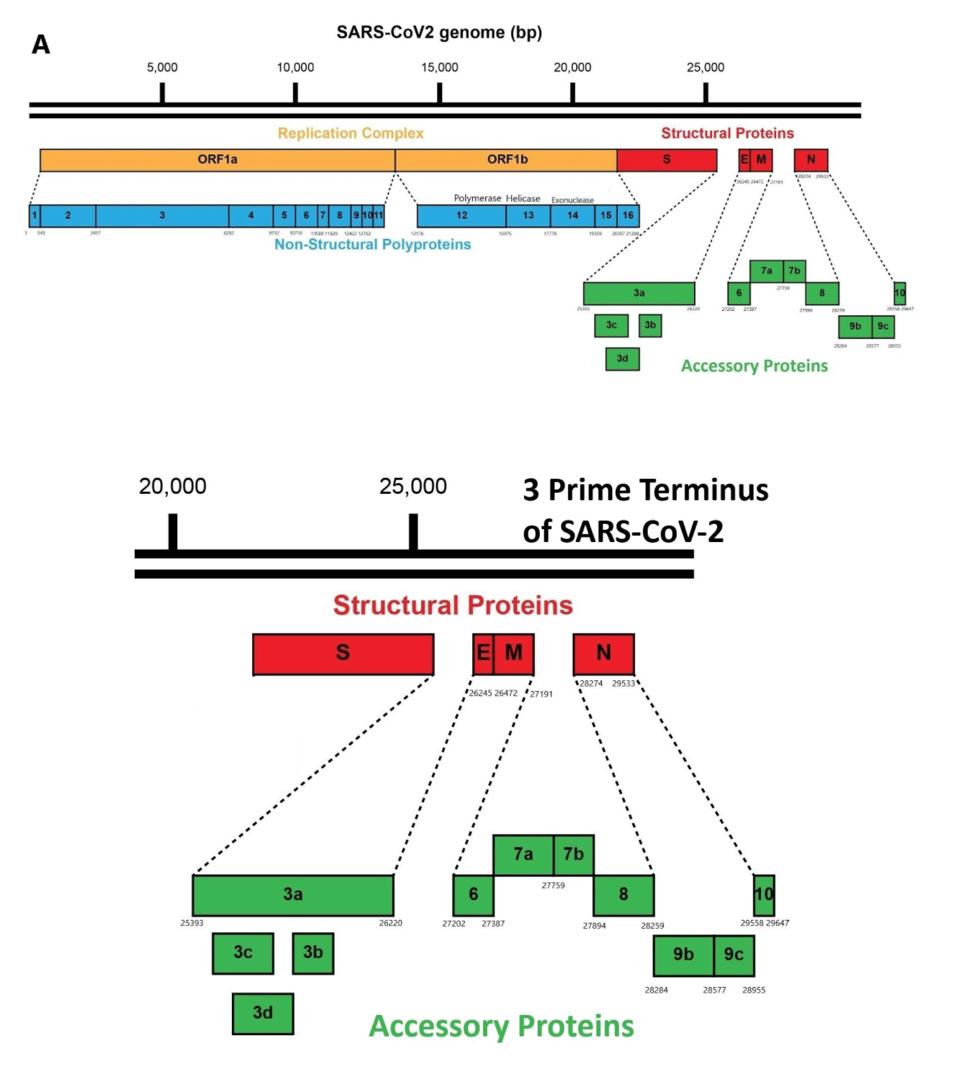
Figure 2: (A) The SARS-CoV-2 genome with each protein labeled, including NSP1-16, S, E, M, N, and Orf3a-10; (B) An expanded view of the SARS-CoV-2 3 prime end.
ACCESS HEALTH INTERNATIONALSARS-CoV-2 encodes 30 proteins. A set of 16 proteins, designated as the nonstructural genes NSP1-16, are encoded by the 5 prime two-thirds of the genome. Upon infection, the full-length viral RNA—which closely resembles a cellular messenger RNA with a 5 prime cap and a polyadenylate tail—serves as the template for the synthesis of these 16 proteins. Together these early proteins create the intracellular conditions for replication and transcription.
The NSP proteins are originally made as long polypeptides, with Orf1a encoding proteins NSP1-10 and an extension, Orf1ab, encoding all 16 NSPs. The individual NSP proteins are cleaved from these precursors by the actions of two proteases, one specified by NSP3 and others by NSP5.
The virus also encodes the four proteins of the infectious virus particle: the S protein that forms the surface spike, the E and M proteins embedded in the viral membrane, and N, the nucleocapsid protein, that forms a complex with the viral genome (Figure 3).
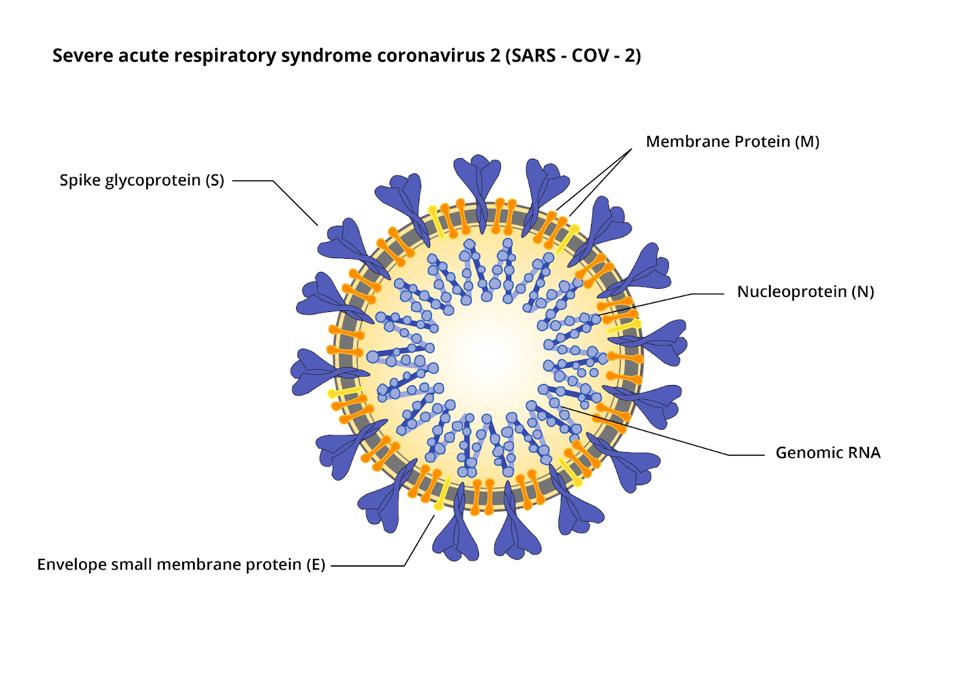
Figure 3: SARS-CoV-2 particle
ACCESS HEALTH INTERNATIONAL; ENCYCLOPEDIA BRITANNICA, INC. HTTPS://WWW.BRITANNICA.COM/SCIENCE/CORONAVIRUS-VIRUS-GROUPInterspersed among the structural genes are the accessory genes designed as open reading frame proteins 3-10 (Orfs 3-10).
SARS-CoV-2 is a member of the nidovirus family, so-called because of their unique means of producing 3 prime terminal messenger RNAs. The messenger RNAs of structural proteins and 3 prime Orf proteins are made from a nested set of transcripts. The 3 prime messenger RNAs are made from subgenomic fragments, each of which serves as a separate replicon, resembling mini-genomes.
Messenger RNA synthesis of the 3 prime genes begins with transcription of the positive genome, creating a nested set of negative strand partial copies that contain both the original 5 prime and 3 prime untranslated terminal sequences. Transcription of a minus strand messenger RNA temple is truncated at the transcription regulation sequences (TRS), followed by a jump to the 5 prime untranslated region. The minus strand is then copied to produce a positive strand. Each subgenomic replicon serves as a template to produce positive-strand messenger RNAs via a process akin to the production of full-length genomic RNA (Figure 4).
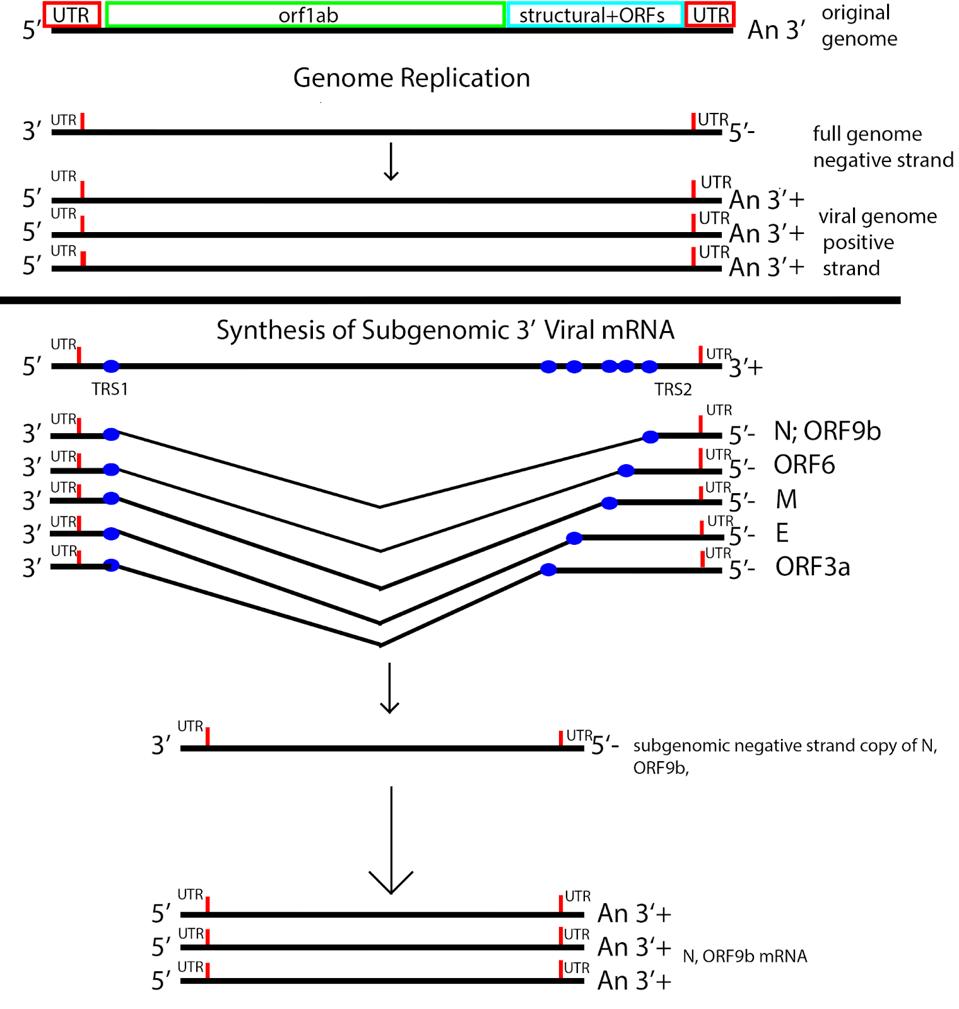
Figure 4: The messenger RNA replication and transcription strategy of SARS-CoV-2. Note the nested set of 3 prime messenger RNAs made by jumping from the 3 prime to the 5 prime transcriptional regulatory sequence.
ACCESS HEALTH INTERNATIONAL; HTTPS://WWW.MDPI.COM/2077-0383/9/4/1225/HTMWe must understand the large complex nature of the viral genome and proteins both for understanding the disease and how to thwart it. As will become clear, many of the viral genes contribute to the virus’s ability to evade the immune system early on, allowing the virus to enter and escape before detection. Disease that occurs two to three weeks later is likely due to dysregulation of the immune system that occurs earlier.
Detailed knowledge of exactly how the virus functions is essential for the development of antiviral drugs that can be used alone or in combination to prevent and treat infections. We are still at an early stage in our understanding. We only know enough to know that we need to know much more. We are in desperate need of a well-funded, highly focused global effort to develop the detailed knowledge we need to create the next set of safe and effective antiviral drugs.
The complex genome and replication strategy of the virus also suggest there are many ways the virus can respond to immune pressure and public health containment strategies. Current efforts focus almost exclusively on interpreting viral variation in terms of changes to the function of the spike protection. Evidence is beginning to emerge suggesting that increases in replication efficiency and immune suppression also contribute to the increase in transmission and virulence of SARS-CoV-2 variants. This unique means of messenger RNA production may provide a selective advantage over many other RNA viruses that must produce all their viral proteins from a single messenger RNA.
I speculate that this strategy allows the virus to preferentially amplify the 3 prime messenger RNAs in response to selective pressure, as recently documented for the Alpha variant. As we shall see in the next piece in this series, preferential messenger RNA amplification contributes to increased immune suppression by the Alpha variant.
Originally published on Forbes (August 16, 2021)

