How SARS-CoV-2 Evades And Suppresses The Immune System (Part 6)
(Posted on Tuesday, August 24, 2021)
This is the sixth article in a 15-part series called “Relevance of Immune Suppression by SARS-CoV-2 to Understanding and Controlling the Covid-19 Pandemic,” which will explore an underappreciated but highly significant aspect of SARS-CoV-2 replication. The ability of SARS-CoV-2 to delay, evade, and suppress the immune system has myriad implications for drugs, vaccines, and other aspects of our pandemic response. The first set of pieces in this series are intended for a general audience; the second set, for the medical community; and the third and final set, for biomedical researchers looking for a deeper understanding of variants, how they’re generated, and what we might do to control them. Read part one, part two, part three, part four, and part five.

Couples at the masquerade ball. You might also be interested in these:
GETTY
In the most recent installment of this series, I described how SARS-CoV-2 evades immune sensors by constructing a secret compartment, a double membrane vesicle, for replication and messenger RNA synthesis. But not only does the virus have to conceal their synthesis, it must camouflage the messenger RNAs themselves so they can continue to evade detection beyond the confines of the compartment. To be translated by the ribosomes, messenger RNAs must leave the compartment. If they leave unprotected, they will trigger the immune sensors.
Normally a class of innate immune receptors called Toll receptors would immediately pick up on the presence of messenger RNA that wasn’t cellular, either by recognizing the naked 5 prime end or lack of methylation, or absence of a polyadenylate 3 prime terminus. Detection by such receptors is followed by activation of secondary messages that in turn activate type-I interferons, which stimulate internal cellular pathways which destroy the RNA and that alert nearby cells to the presence of an invader. By masquerading its genome and messenger RNAs as cellular, SARS-CoV-2 produces enough virus to escape and infect other human hosts before immune defenses are fully mobilized.

Costume Ball at the Opera – after engraving by Jean Francois Bosio after 1800. Paris. With harlequin figure in centre (Commedia dell’arte character). (Photo by Culture Club/Getty Images) *** Local Caption ***
GETTY IMAGESFor all the privileges double membrane vesicles afford SARS-CoV-2, they do exclude ribosomes, a piece of cellular machinery the virus needs to make the proteins required for successful replication. For that reason, messenger RNAs must be released into the cytoplasm. And they too must closely resemble cellular messenger RNAs.
Synthesis of proteins required for replication
As it enters the cell, the viral genome in effect serves as the messenger RNA for synthesis of the first set of proteins required for replication. These proteins are specified by the initial two thirds of the viral genome. They are encoded by two long open reading frames, designated Orf1a and Orf1b. They are translated as a full length Orf1a protein and an Orf1a1b protein. Elongation of the Orf1a into the Orf1ab protein occurs by a -1 frameshift as the ribosomes translate the viral genome.
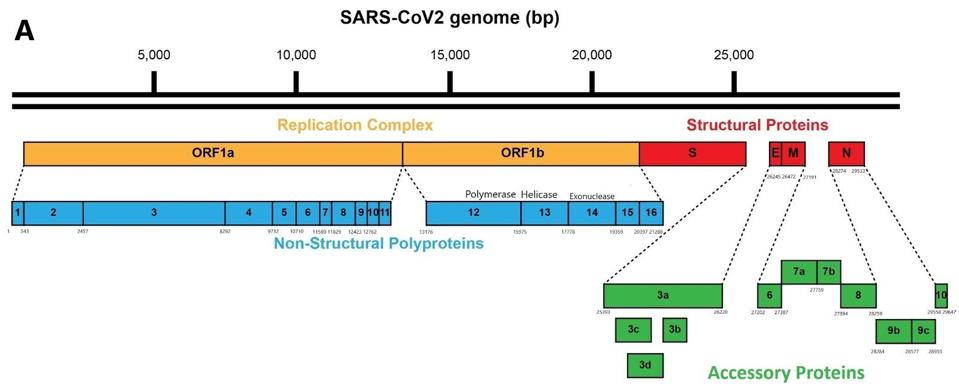
Figure 1: Schematic of the SARS-CoV-2 viral genome.
ACCESS HEALTH INTERNATIONAL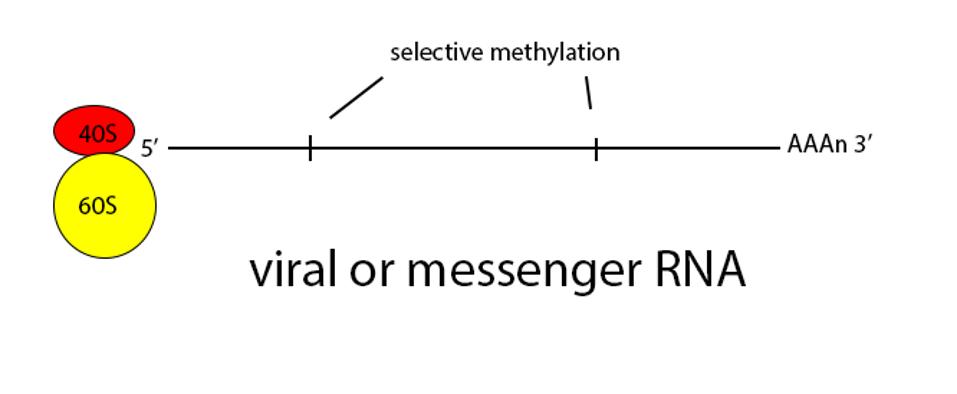
Figure 2: Schematic diagram of translation.
ACCESS HEALTH INTERNATIONAL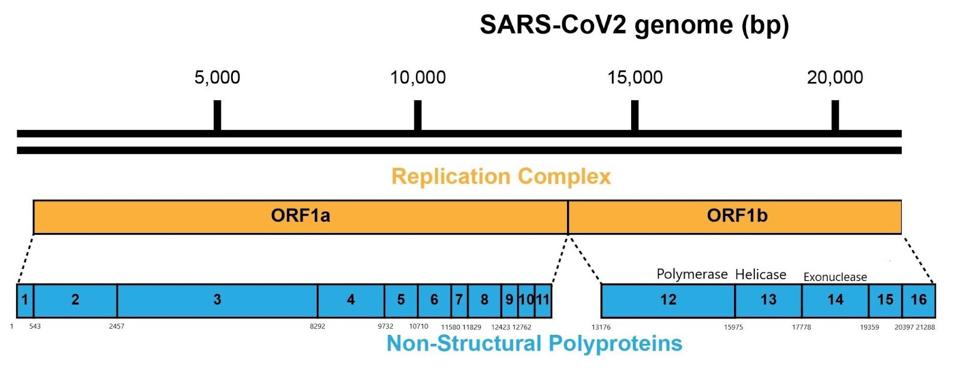
Figure 3: A schematic representation of the SARS-CoV-2 genome and polyproteins Orf1a and Orf1b.
ACCESS HEALTH INTERNATIONALSome of these proteins help create the special compartment. Four of these replication proteins, NSP10, NSP13, NSP14, and NSP16, add the 5 prime cap and methylation so they closely resemble messenger RNAs. The genome also comes equipped with a polyadenylate tail, as do all messenger RNAs.
The formation of the complex requires a combination of enzymes that use cellular constituents to build a cap. Cellular mRNAs have the cap put on in the nucleus, where they are transcribed. However, the viral genome never reaches the cell nucleus. Therefore the cap must be attached during the process of genomic synthesis. That requires a minimum of four virally encoded enzymes. These enzymes are among the first made by translation of Orf1a. One is made from Orf1a, NSP10, and the remaining three are made from Orf1a1b.
The next step is producing 3 prime messenger RNAs. To do this SARS-CoV-2 uses a methodology unique to its family of viruses, Nidovirales, by which 3 prime messenger RNAs are made beginning with transcription of the positive genome, creating a nested set of negative strand partial copies that contain both the original 5 prime and 3 prime untranslated terminal sequences.
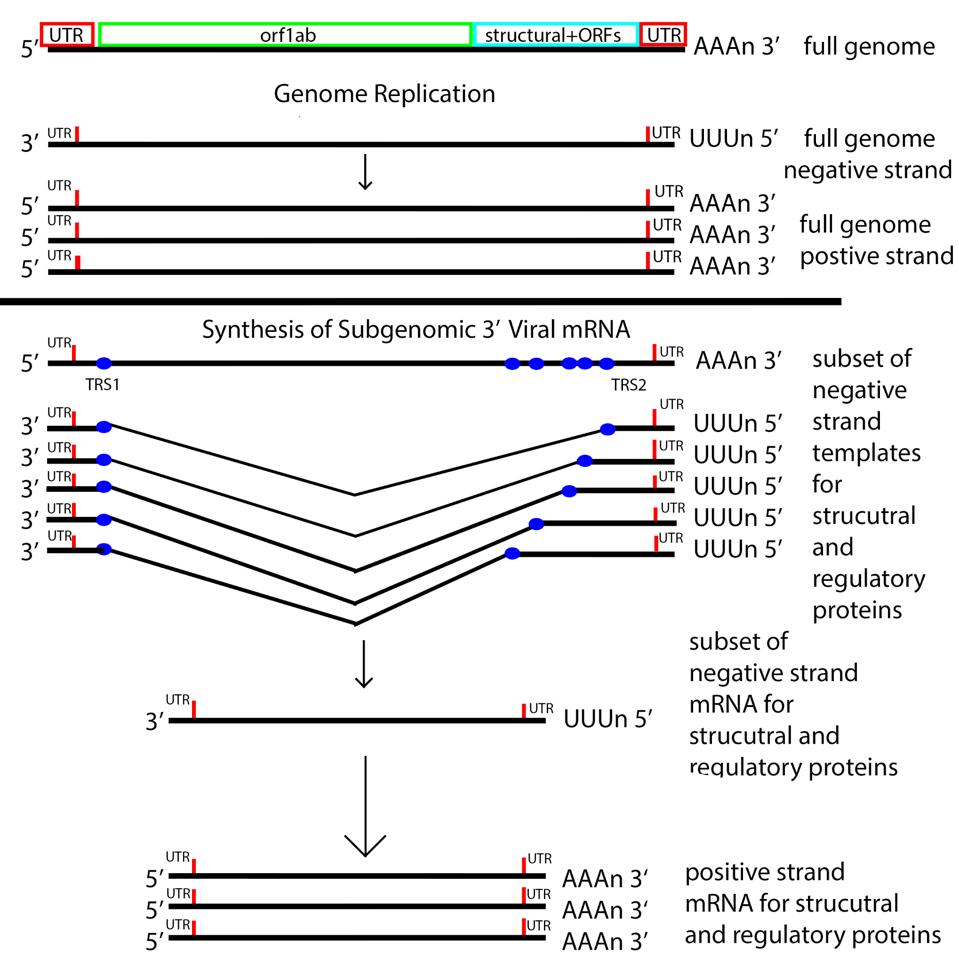
Figure 5: The messenger RNA replication and transcription strategy of SARS-CoV-2. Note the nested set of 3 prime messenger RNAs made by jumping from the 3 prime to the 5 prime transcriptional regulatory sequence.
ACCESS HEALTH INTERNATIONALOnce viral messenger RNAs exit the compartment through the specialized molecular pore into the cytoplasm for translation, they’re methylated with a cap at the 5 prime end. Presumably the polyadenylate tail is derived from the original genomic polyadenylate tail. The primary enzyme that adds polyadenylates to nascent cellular messenger RNAs is found in the nucleus. However, there is a second polyadenylation enzyme present in the cytoplasm which may add additional adenylate residues to the 3 prime end of the messenger RNA. Collectively these processes thoroughly disguise the viral messenger RNAs as cellular.
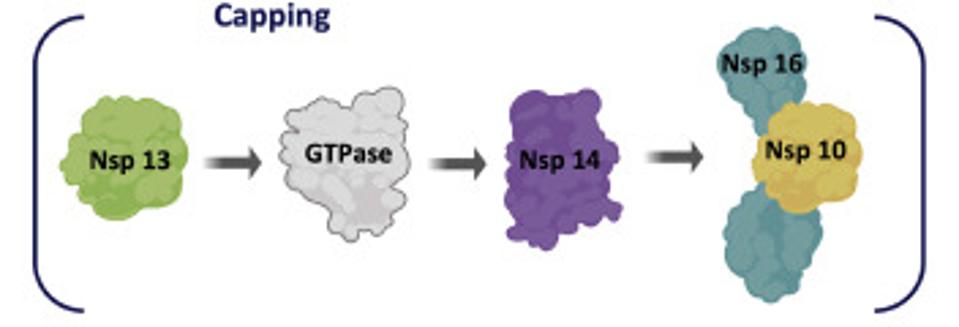
Figure 4: Nonstructural proteins that participate in formation of the cap and methylation complex.
ACCESS HEALTH INTERNATIONAL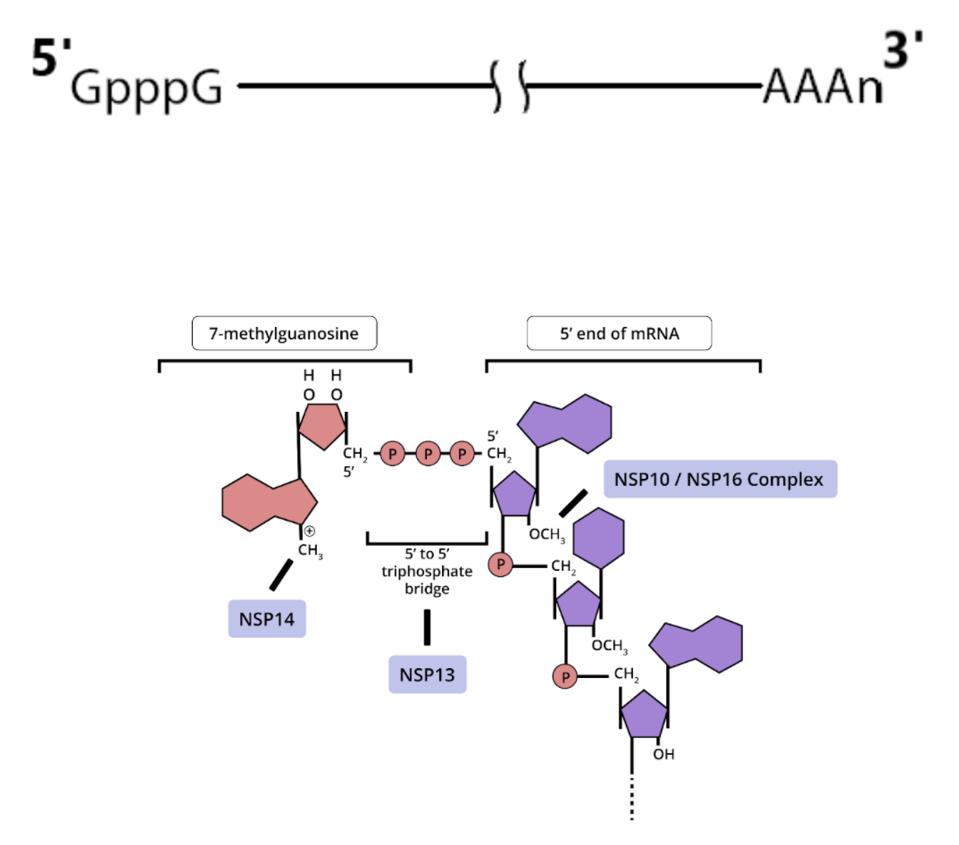
Figure 5: (A) Also involved in this process are the RNA/NTP triphosphatase (TPase) and helicase activity, which is part of the NSP13 protein. NSP13 is essential for cap formation, breaking down nascent RNA so a guanosine monophosphate moiety can
ACCESS HEALTH INTERNATIONALFortunately the three-dimensional structure of each of these proteins is known in great detail and forms the basis for finding small molecule drugs that will interfere with viral activity. Any one of these, used alone or in combination, should be sufficient to inhibit the virus. Their discovery will bring us significantly closer to the day when we’ll have multimodal Covid-19 therapies to prevent infection and disease.
In my next piece for this series I will discuss how SARS-CoV-2 promotes selective translation of messenger RNAs through the nonstructural protein NSP1.
Originally published on Forbes (August 23, 2021)

