Decoding The Gordian Knots At The Ends Of The SARS-CoV-2 Genome
(Posted on Wednesday, August 25, 2021)
This is the first in a series describing the role of the beginning and end of the SARS-CoV-2 genome in the virus life cycle. I summarize what we know and point out what we need to know about these ends in order to develop new antiviral drugs.

Alexander the Great Cutting The Gordian Knot
FEDELE FISCHETTI
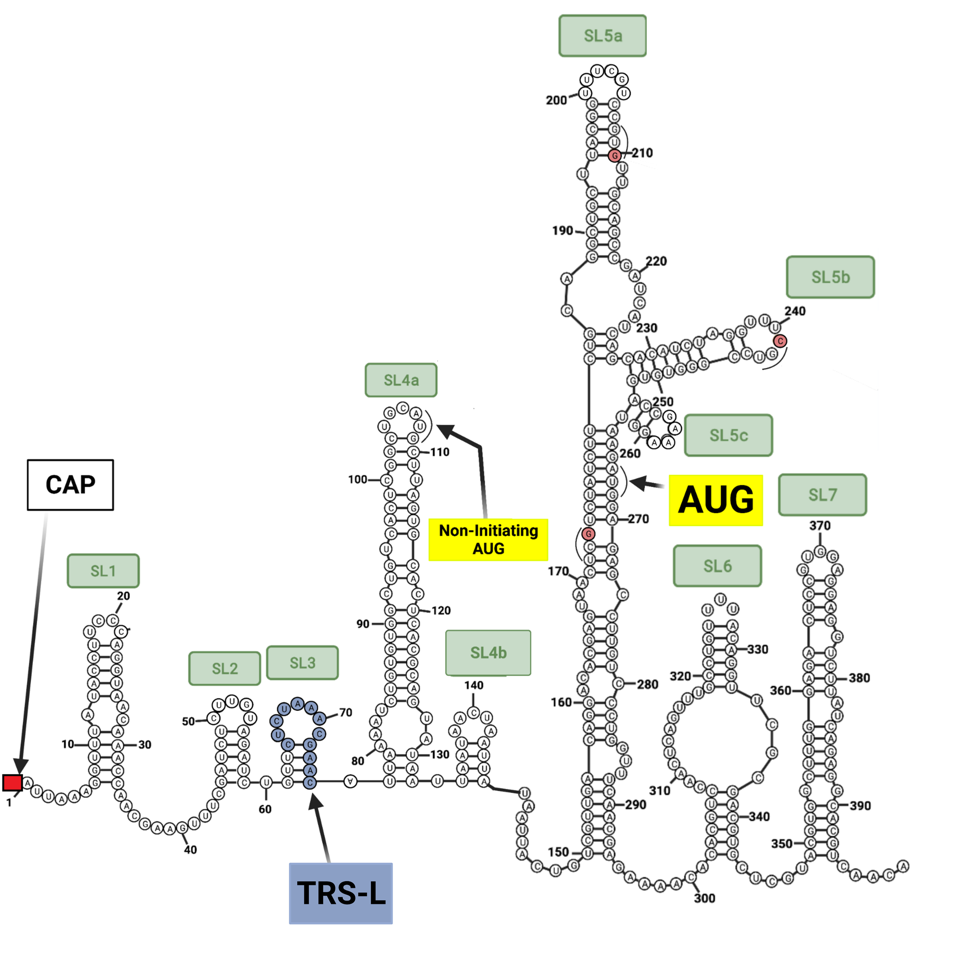
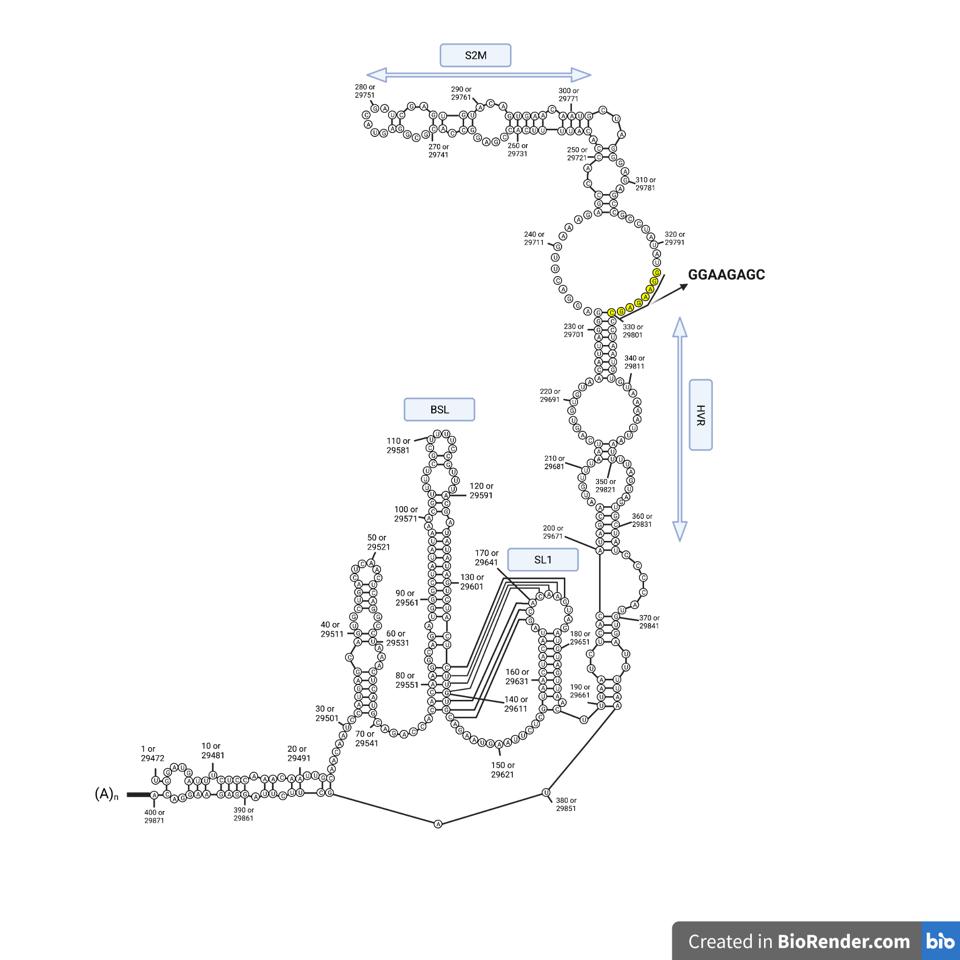
Unraveling the details of the life cycle of the SARS-CoV-2 virus is much like reading a mystery novel. The truth is often deeply buried and the journey laden with misleading clues. As every reader knows, the first and last pages are inevitably the most important. The analogy is closer than you may imagine. The bookends of the viral genome are responsible for many of the virus’s critical functions, including initiation of replication, protein synthesis, and messenger RNA synthesis. Unraveling the details of exactly how these functions occur requires puzzling out the most intricate mysteries. The first observation is that both the beginning five prime (5’) end and terminal three prime (3’) end are complex (Figure 1).
ACCESS HEALTH INTERNATIONAL
ACCESS HEALTH INTERNATIONAL

FIGURE 1: (A) 5’ end of the SARS-CoV-2 genome; (B) 3’ end of the SARS-CoV-2 genome; (C) Gordian Knot
GORDIAN KNOT
RNA is a self-folding polymer. The 30,000-nucleotide long genome self-assembles into elaborate stem-loop structures. The stems are base-paired, G pairs with C and A with U (or T in the case of DNA), while the loops are unpaired (Figure 2).
MARKOVA ET AL
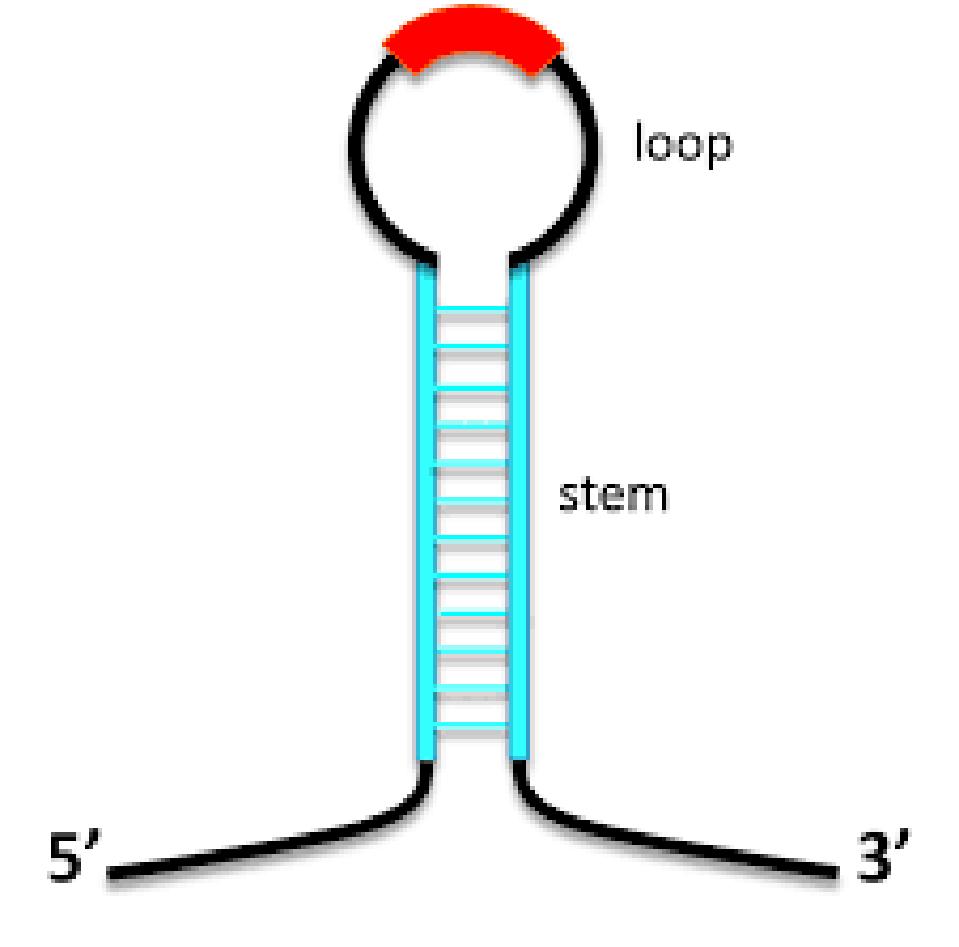
FIGURE 2: (A) Theoretical stem-loop structure; (B) Schematic representation of a stem-loop.
IGEM.ORG
Alternatively, these structures can fold into pseudoknots, emerging in various forms (Figure 3).
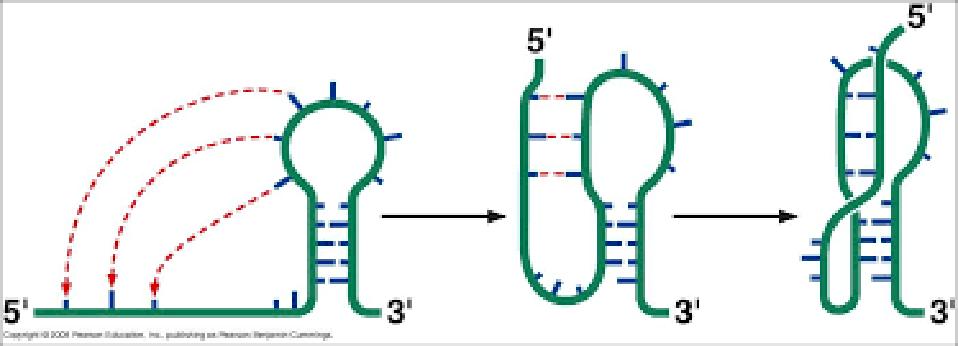
FIGURE 3: Theoretical pseudoknot stem-loop structures
RUTGERS UNIVERSITY
Collectively, these are called secondary structures. The secondary structures of diverse coronaviruses are relatively well conserved, even though the primary genome sequences differ. The similarity of these structures suggests that the structure itself, in addition to the primary nucleotide sequence, plays an essential role in virus replication. Moreover, understanding the details of how the ends of the viral genome interact with viral and cellular proteins is a prerequisite to the discovery of new antiviral drugs.
The recent introduction of a new drug to prevent and treat influenza illustrates exactly how important this research is in finding new drugs to prevent and treat SARS-CoV-2 infection. A study from July 2020 found that the anti-influenza drug baloxavir marboxil (also referred to as Xofluza), reduced close-quarters transmission of Influenza A by 80%.
The influenza virus uses a process called cap snatching to reproduce, effectively snatching host RNA and reusing it in the reproduction process. Xofluza binds to and inhibits the proteins involved in the cap snatching process.
My hope is that these brief descriptions of what we know and don’t know about the termini of SARS-CoV-2 will stimulate scientists around the world to develop drugs similar to Xofluza for the virus at hand.
Entry of the viral genome and production of the viral replication complex
Upon fusion of the viral and cellular membrane, the viral genome is deposited into the cytoplasm of the cell. The first requirement of the viral RNA is to avoid triggering the antiviral defenses, collectively called the innate immune response. A primary trigger of the innate immune response is the entry of foreign RNA. The cellular alarm signals recognize naked RNA 5’ termini, unmethylated RNA, and RNA that does not carry a polyadenylated (poly-A tail). The SARS-CoV-2 genomic RNA skirts all these alarm signals as it is properly capped and methylated by the virus’s own proteins. It also carries a poly-A tail. In other words, it masquerades as a cellular messenger RNA.
Once safely inside the cytoplasm, replication begins. As it enters a cell, the viral genome is organized as a compact package bound to multiple nucleocapsid capsid (N) proteins (Figure 4). The viral genome itself serves as a template for the synthesis of the very first viral proteins located in a long open reading frame that begins at the “AUG” initiation codon located 266 nucleotides from the 5 prime end of the genome, buried deep within the 5’ stem-loop structures.
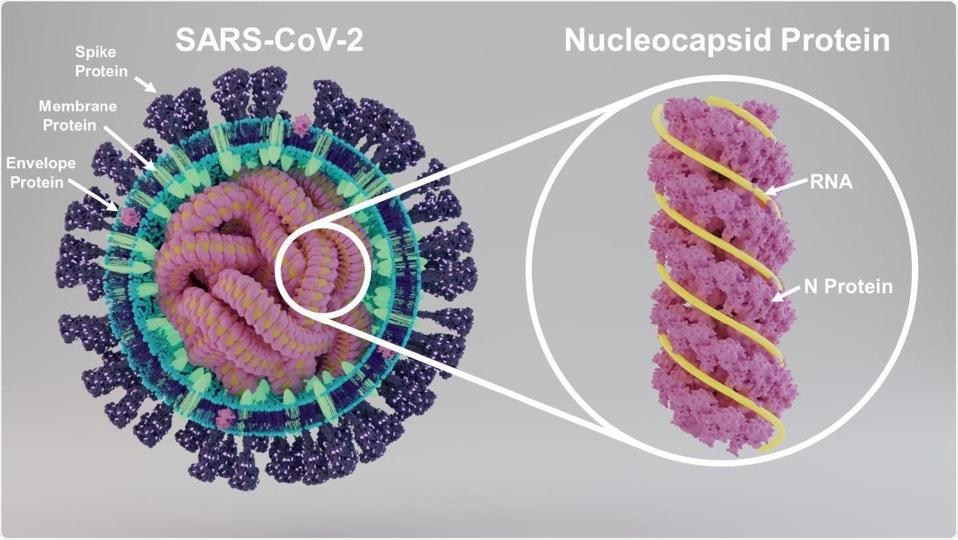
FIGURE 4: Viral RNA bound to the N protein.
LIJI THOMAS / NEWS MEDICAL
Question: How do the translation machinery, translation initiation, and associated initiation factors recognize the genome RNA complexed with the N protein. Does the N protein disassociate from the RNA on entry spontaneously or is it displaced by cellular proteins and the ribosome during protein synthesis?
Virus protein synthesis begins when the ribosomes bind the 5′ end of the genome and initiate synthesis (Figure 5).
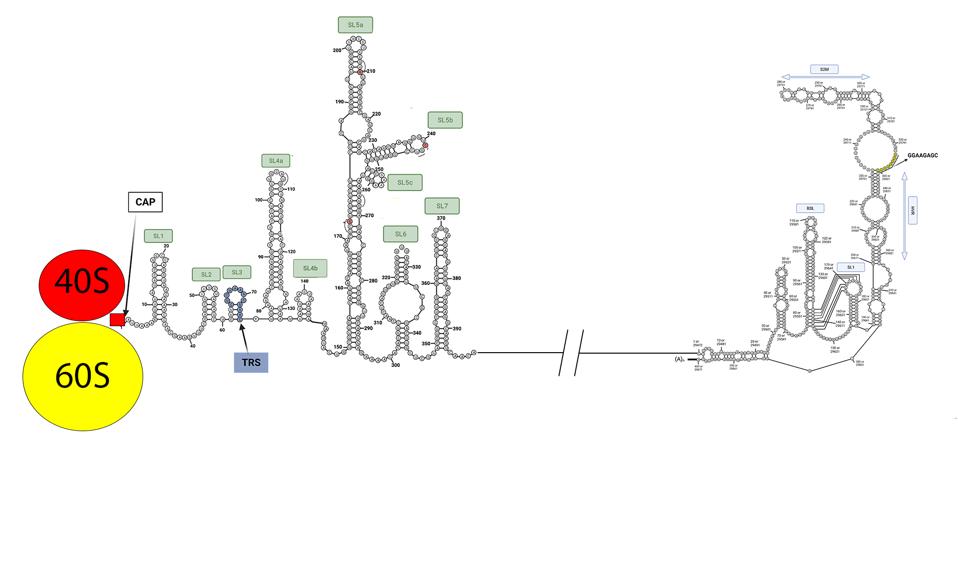
FIGURE 5: Messenger RNA being translated by a ribosome.
ACCESS HEALTH INTERNATIONAL
It is no mean feat for the ribosome to navigate the intricate 5’ structure. Ribosomal entry likely requires the assistance of a cellular unwinding enzyme (a helicase) allowing slippage of the ribosome along the RNA until it encounters the “AUG” initiation codon. Initiation of the Orf1a and Orf1b proteins begins at position 266 with stem-loop 5 (Figure 1).
We note that there is the possibility of initiation at position 107. There is an “AUG” codon within the reading frame at this position which would proceed to yield a nine-amino acid long peptide before encountering two termination codons
I wonder if initiation at position 107 followed by reinitiation at position 266 is possible. A similar 5’ “AUG” occurs in SARS-CoV. As the structure of the stem-loops is nearly identical and the resulting theoretical peptide in SARS-CoV from the first “AUG” closely resembles that of SARS-CoV-2, this is not an anomaly confined to SARS-CoV-2 (Figure 6). It would be interesting to learn if this small peptide is functional and we will continue to research this sequence in an effort to elucidate it.
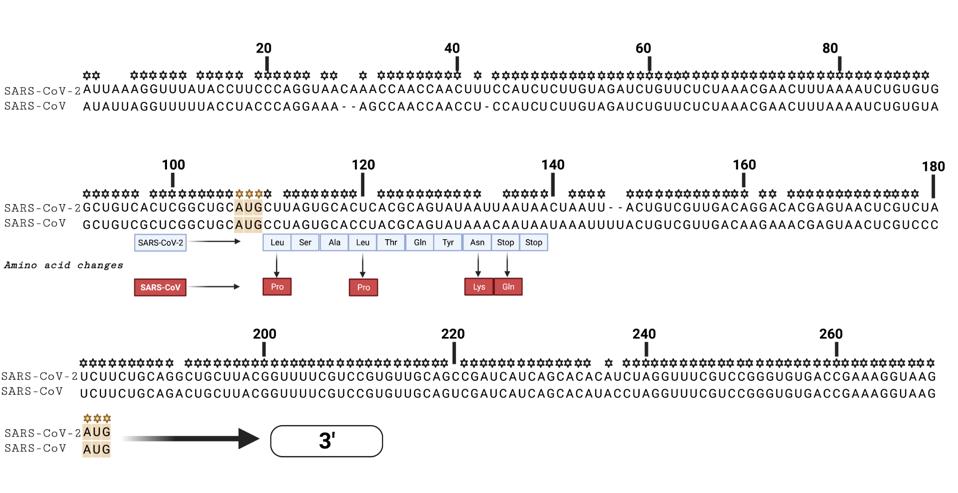
FIGURE 6: SARS-CoV and SARS-CoV-2 5’ ends, denoting the initial “AUG” codon that results in a nine amino acid long peptide before the Orf1ab initiating “AUG” downstream in the sequence. Note the resultant amino acid peptides in both SARS-CoV and
ACCESS HEALTH INTERNATIONAL
Question: Why does Orf1a protein start at the second “AUG” at position 266 and not the first “AUG” at position 107 (Figure 1). The general rule is that protein synthesis in mammalian cells begins at the initiation codon closest to the 5’ end. Is it possible that initiation actually begins at position 107 to yield a nine amino acid-long peptide of unknown function followed by re-initiation at position 266?
The first proteins made are the two products of the Orf1a and Orf1b genes. These long polypeptides are cleaved into 15 proteins, called the non-structural proteins (NSPs1-16: there is no NSP11) that are required for the synthesis of small messenger RNA that direct the synthesis of the proteins of the virus particle S, M. E, and N, as well as the regulatory proteins.
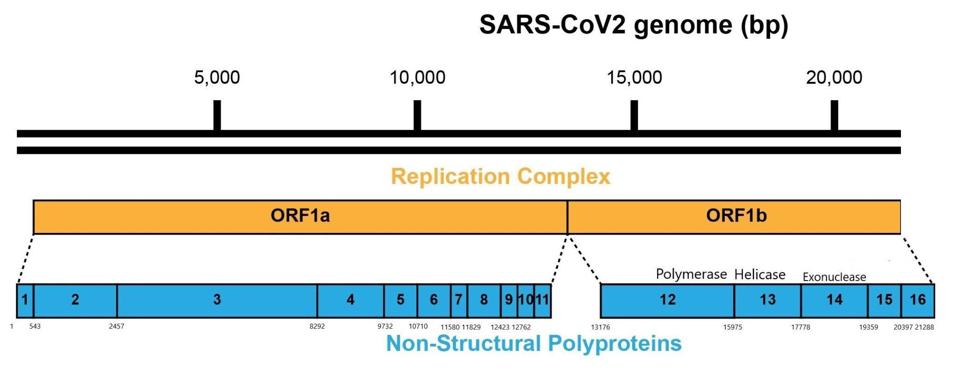
FIGURE 7: Frameshift between Orf1a and Orf1b
ACCESS HEALTH INTERNATIONAL
The very first viral protein made, NSP1, plays a critical role in virus replication. The protein blocks the production of cellular proteins while permitting the production of viral proteins. NSP1 acts by obstructing the entry of cellular RNAs into the ribosome (Figure 8).
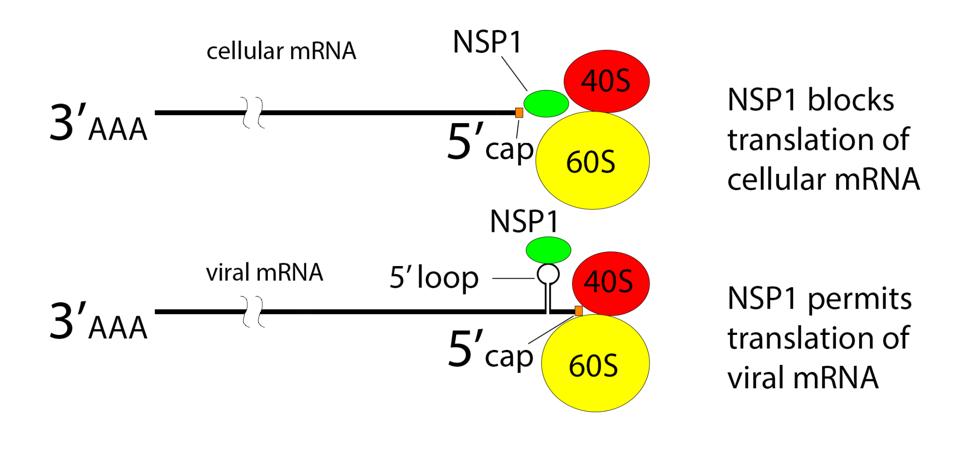
FIGURE 8: Ribosomal translation
ACCESS HEALTH INTERNATIONAL
Recent studies show that the N protein binds to an RNA sequence of the 40S subunit of the ribosome obstructing the entry tunnel. Preferential synthesis of viral proteins allows much of the cell’s energy to be devoted to producing viral components.
How then are viral proteins made? The mystery was solved by studies that show any message with stem-loop 1 located near the 5’ terminus can be translated in the presence of NSP1. Both the viral genome and all viral messenger RNAs meet this requirement.
A brief description of viral messenger RNA synthesis explains why all viral messages carry the requisite 5’ stem-loop. The structure protein S, M, E, and N of the virus particle and regulatory genes Orfs by the 3’ end of the genome. The template for their synthesis is a nested set of negative-strand RNAs that all share a 3′ and 5′ termini (Figure 9).
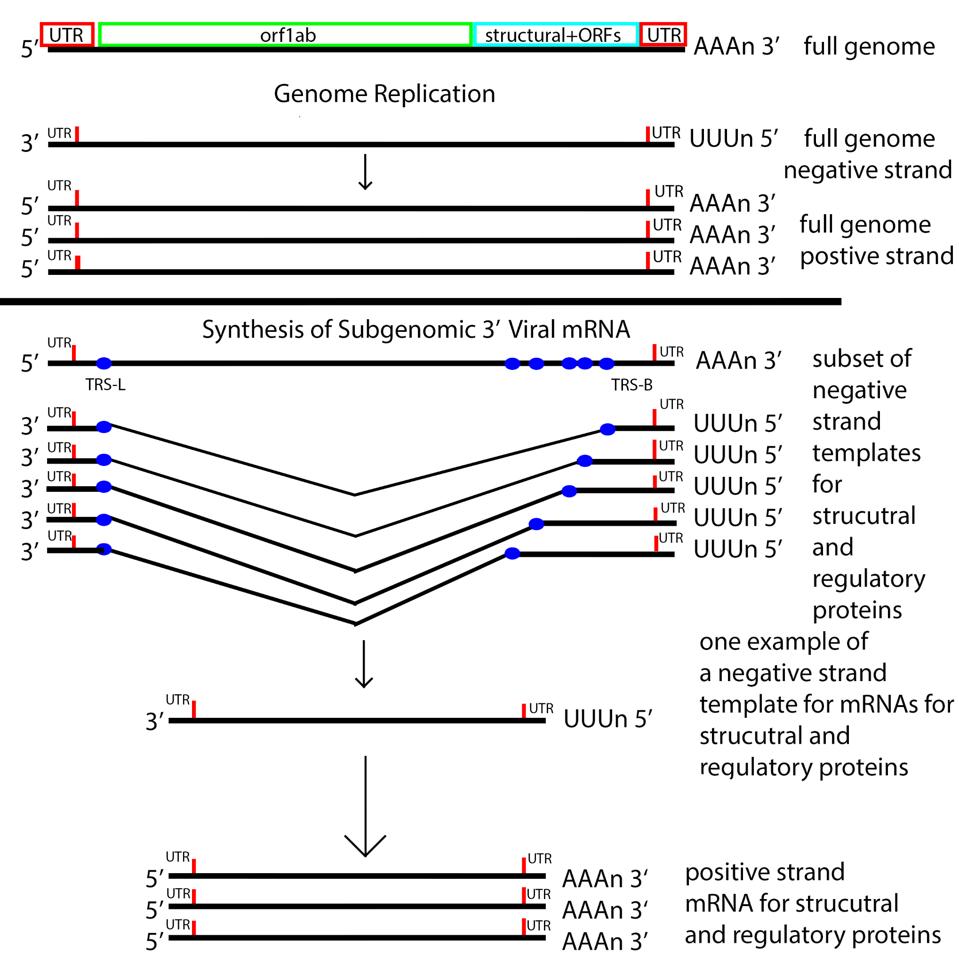
FIGURE 9: The messenger RNA replication and transcription strategy of SARS-CoV-2. Note the nested set of 3’ messenger RNAs made by jumping from the 3’ to the 5’ transcriptional regulatory sequence.
ACCESS HEALTH INTERNATIONAL
As the negative strand elongates, the growing end encounters what’s called termination regulatory sequences (TRS-B). Transcription pauses after copying the TRS sequences (Figure 10) and then resumes by pairing with the complementary TRS-L sequence located near the 5’ terminus.
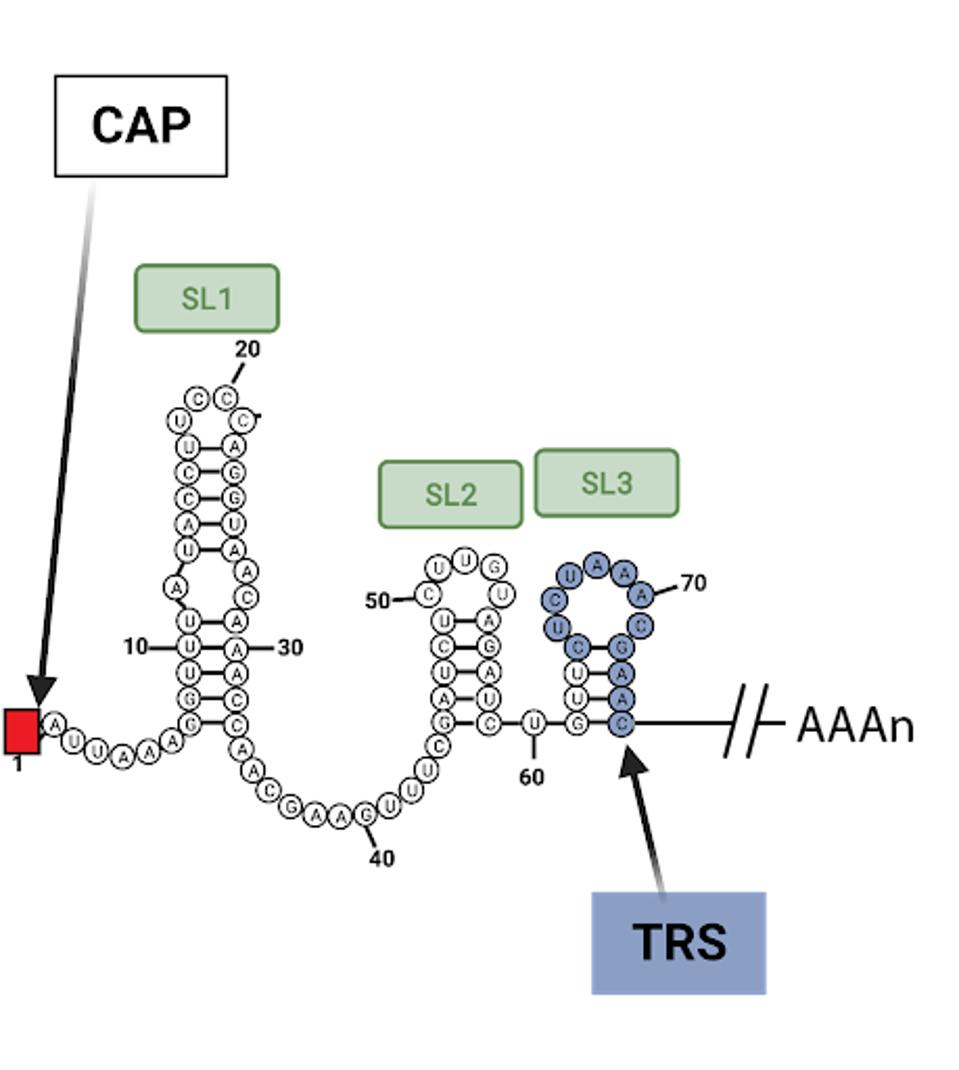
FIGURE 10: 5’ End through TRS-L at nucleotide position 75.
ACCESS HEALTH INTERNATIONAL
The messenger RNAs all begin with the same 5 ’end that extends from nucleotide 1 through nucleotide 75 and includes stem-loops 1-3, but not stem-loops 4-7.
Question: How does the presence of stem-loop 1 relieve the NSP1 translation block? Does NSP1 bind to stem-loop 1? If so is NSP1 recognition of stem-loop 1 determined by the sequence or structure. Can stem-loop 1 of SARS-CoV-2 relieve the NSP1 block of other coronaviruses? What is the role of cellular translation initiation factors in viral messenger RNA translation?
Question: Why does transcription pause at TRS-B sequences. Does the topology of the replication complex facilitate the post pause jump to the 5’ TRS? Can the jump occur only in cis to the same genome or is a trans jump possible to a second replicating genome? Can a jump occur to any TRS sequence or only the one closest to the TRS-L sequence 5’ end? Do cellular proteins participate in messenger RNA synthesis?
Read the full article on Forbes, (originally published August 25th, 2021.)

