The Mystery Of The False Start At The 5’ End Of SARS-CoV-2
(Posted on Monday, August 30, 2021)
This is the second in a series describing the role of the beginning and ends of the SARS-CoV-2 genome in the virus life cycle. I summarize what we know and point out what we need to know about these ends in order to develop new antiviral drugs. Read more from this series in part one.

Mauritania’s Hamza Labeid (front) dives prior to the start of the heats of the men’s 50-metre freestyle swimming event in the FINA World Championships at the indoor stadium of the Oriental Sports Center in Shanghai on July 29, 2011. Labeid was
AFP VIA GETTY IMAGES
In every mystery story, there are unsolved questions, loose ends, and false leads. Here we discuss an anomaly that may be either an important clue in SARS-CoV-2 replication and pathogenesis or may turn out to be irrelevant. There is a short string of nucleotides in SARS-CoV-2, as well as most coronaviruses, that has the potential to initiate protein synthesis between 150 and 200 nucleotides prior to the initiation of the first major protein in the virus, Orf1a.
To recall, protein synthesis beings with the codon “AUG.” This codon specifies the amino acid methionine. Elongation of a protein continues as the translation machinery reads the nucleic acid sequence in groups of three until it terminates at well-defined groups of three nucleotides called stop codons. The stop codon comes in three forms: “UAA,” “UAG,” and “UGA.”
We have observed that for SARS-CoV-2, there is an “AUG” in stem-loop four at position 107, preceding the “AUG” in stem-loop five at nucleotide position 266, which is the initiation codon used for the precursor proteins Orf1a and Orf1ab. The stem-loop four “AUG” codon and following nucleotide sequence could potentially encode a peptide nine amino acids long in SARS-CoV-2 between positions 107 and 133 (Figure 1).

FIGURE 1: SARS-CoV-2 5’ end secondary structure with both the uOrf and Orf1a initiating “AUG” codons highlighted. The red box indicates stem-loop four, where the uOrf and uOrf Kozak sequence are contained.
ACCESS HEALTH INTERNATIONAL
We note, as have others, that many other coronaviruses contain an “AUG” 5’ to the Orf1a initiation codon in a similar position within the stem-loop structure, such as mouse coronavirus (MHV), bovine coronavirus (BCoV), MERS-CoV, and SARS-CoV-1 (Figure 2). Many of these also have the potential to code short peptide sequences ranging in length from three to thirteen amino acids. These sequences are not well conserved.

FIGURE 2: Comparison of secondary structure models of the 5′ regions of MHV-A59, BCoV, MERS-CoV and SARS-CoV-1.
YANG AND LEIBOWITZ
The figure below illustrates the similarities and differences of the peptide sequences. We note that SARS-CoV-1 and SARS-CoV-2 are similarly terminated with double stops. Additionally, six of the nine amino acids are identical, indicating some conservation between SARS-CoV-1 and SARS-CoV-2.
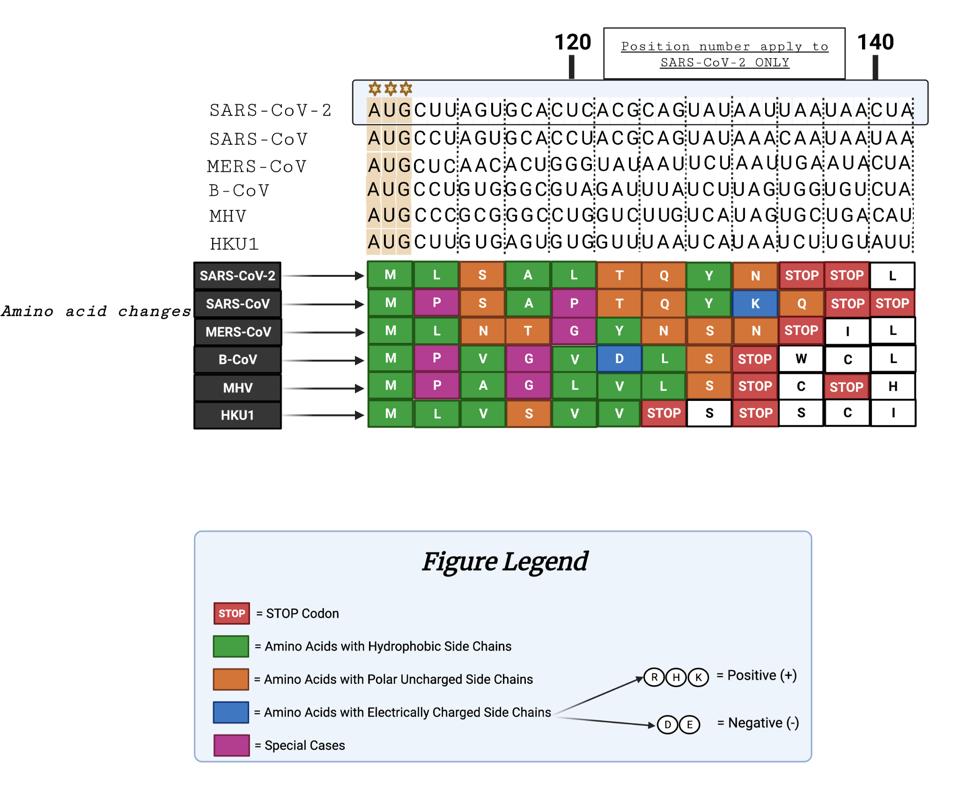
FIGURE 3: Comparison of coronavirus 5’ uOrfs. From top to bottom: nucleotide sequences following the 5’ “AUG” codon for varying coronaviruses; resultant amino acids from nucleotide sequences; figure legend.
ACCESS HEALTH INTERNATIONAL
Hung-Yi et al indicate that more than 75% of coronavirus genomes share the characteristic AUG-initiated 5’ uOrf, indicating remarkably high cross-species conservation. For the remaining 25%, Hung-Yi and colleagues note an anomalous potential “CUG” codon-initiated open reading frame.
These sequences are ultimately defined in a larger context by the Kozak sequence, which is a nucleic acid motif that facilitates ribosomal initiation in eukaryotic cells.
The optimal Kozak sequence is GCCGCC(A or G)CCAUGG. The RNA sequence in question does not need to match the Kozak sequence exactly, but higher similarity correlates to greater translation success. The sequence leading up to the 5’ “AUG” of SARS-CoV-2 is CUCGGCUGCAUGC. Although this sequence does not perfectly match the Kozak sequence, it should suffice to allow some level of translation of the SARS-CoV-2 uOrf peptide.
These sequences are relevant only if they are translated into functional polypeptides. Hung-Yi et al investigated this issue with the mouse coronavirus. They observed that the small polypeptide in MHV was synthesized. They also investigated whether the 5’ uOrf was functional. They found that mutations that prevented the synthesis of uOrf but maintained the secondary structure, appeared to be fully viable in cell culture. However, serial passage of these mutant variants resulted in a reversion to the wild type, indicating that the “AUG” does confer a selective advantage in replication in cell culture.
Hung Yi et al suggest that a functional uOrf peptide sequence 5’ to the initiation codon for Orf1a has the potential to decrease ribosomal translation efficiency. The attenuation of Orf1a and Orf1ab synthesis is required for the balanced stoichiometry of virus particle formation. That is the ratio of the 5’ replicative proteins necessary to create the replication complex to 3’ structural and regulatory proteins.
While a full functional analysis of the 5’ uOrf in different coronaviruses remains to be completed, there are some indications that they play some role in replication. A study by Raman et al notes that the stem-loop structure which contains the 5’ uOrf amino acids for BCoV, when deleted, decreases the replication competence of defective interfering particles. This is indirect evidence to suggest that at a minimum, the stem-loop containing the uOrf is positively correlated with viral replication competency.
Missing from the collection of studies on the 5’ uOrf is its role in replication and pathogenesis in animal disease models. This lacuna in our knowledge should be promptly addressed experimentally with MHV, MERS-CoV, SARS-CoV-1, and SARS-CoV-2.
Read the full article on Forbes, (originally published August 20th, 2021).

