Origin: Very Close Relatives Of SARS-CoV-2 Identified In Laotian Bats
(Posted on Wednesday, October 6, 2021)

This horde of bats could contain possible carriers of the rabies virus, 1972. Most of the recent human rabies cases in the United States have been caused by rabies virus that was transmitted through a bat vector. However, it is rare for humans to contract rabies from infected bats.
GETTY IMAGES
The pandemic SARS-CoV-2 virus likely originated from bats in Southeast Asia. There are over 450 million people sharing the region with the bats that live in limestone caves in countries like Vietnam, Thailand, Myanmar, and Laos. One of the predominant genera of bats in the region is the Rhinolophus (horseshoe bat). Bat species falling in this genus contain an ACE2 receptor that closely resembles that of humans. The ACE2 is so similar, in fact, that many of the coronaviruses in these bats have the potential to infect humans as well.
The exact origins of the SARS-CoV-2 virus are shrouded in mystery and some controversy. Recent estimates of the number of people infected by coronaviruses of horseshoe bat origin in Southeast Asia are between 400,000 and two million. A recent study from the Pasteur Institute, Temmam et al., investigates viruses endemic to the bats of this region.
The researchers sampled 645 bats from four different Laotian sites. The bats were classified into 46 different species. It should come as there are over 1,400 different species of bats throughout the world. They extracted the coronaviruses from the bats’ blood, feces, urine, and saliva.
The first finding is that as many as seven different species of bats harbored the same type of coronavirus, known as a sarbecovirus. These are a subgenus of betacoronaviruses, to which the SARS-CoV-2 and SARS-CoV species of virus belong.
In a startling discovery, the authors report that several of the viruses isolated from bats caves in the Vientiane Province are very closely related to SARS-CoV-2. In fact, they are so closely related that the idea that SARS-CoV-2 originated from RaTG13 coronavirus isolated from horseshoe bat in southern China in 2013 is no longer tenable. The receptor-binding domains of SARS-CoV-2 and RaTG13 differ too greatly.
The original SARS-CoV-2 Wuhan strain has 17 amino acid contact points between the virus and the ACE2 receptor.
Several of these are modified in the RaTG13 coronavirus. However, two of the viruses analyzed by Temmam et al.—BANAL-20-52 and BANAL-20-103—differs by only one of the 17 amino acids. Another analyzed virus—BANAL-20-236—differs by only two amino acids. By contrast, in RaTG13, there are six amino acid modifications. We note that a change of only one or two residues in the contact points is well within the range of various naturally occurring variants of SARS-CoV-2, which carry as many as three amino acid changes in these residues.
The authors conducted several experiments to determine whether similarity in the receptor-binding domain region of the Spike protein of the BANAL-20-52, 103, and 236 translated to a similar function to that of SARS-CoV-2. Their first question was whether the receptor-binding domain of the viruses is capable of infecting cells that carry the human ACE2 receptor. Using a pseudotype virus on cells in culture, they found that the answer is a resounding yes. They went on to show that the complete genome of BANAL-20-236 is plaque-forming when introduced to human cells. In fact, it formed plaques more so than the Wuhan strain of SARS-CoV-2. These two observations demonstrate that not only does the bat virus receptor-binding domain recognize human ACE2, but the whole virus is capable of infecting humans.
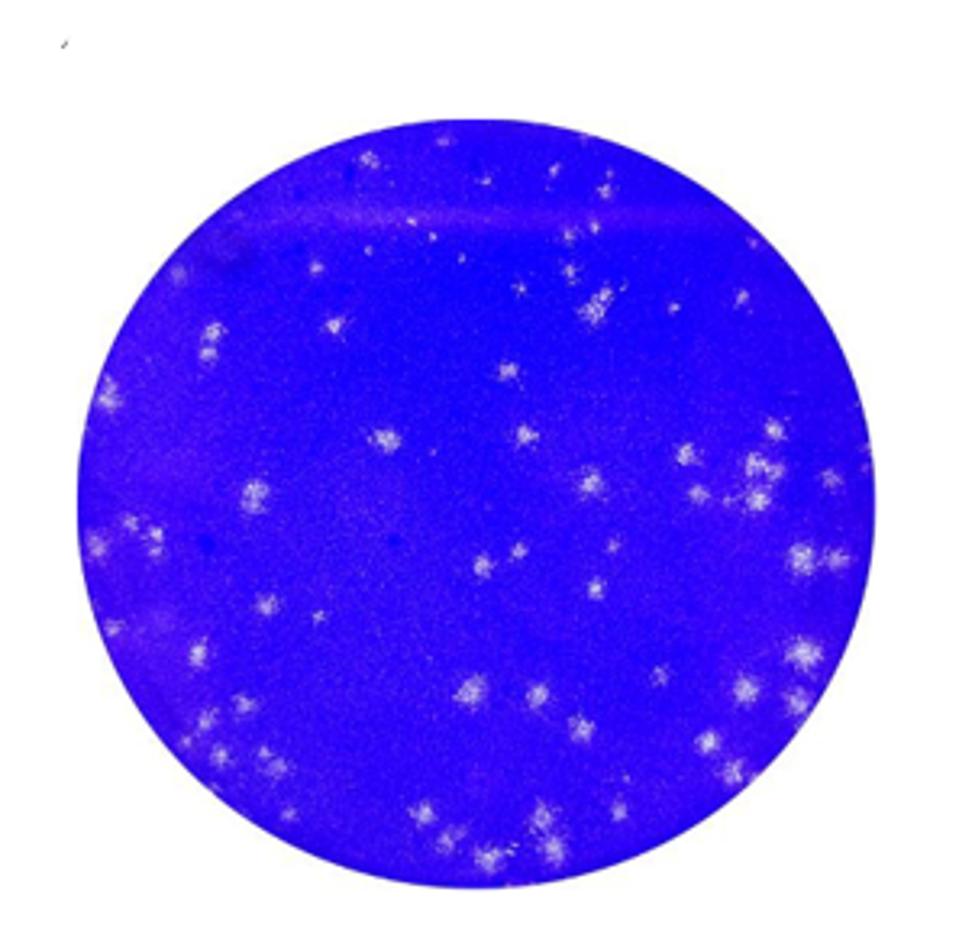
FIGURE 1: Infectivity and Plaques of BANAL-20-236 on human VeroE6. Figure from Temmam et al.
TEMMAM ET AL
Their second concern was to understand exactly how similarly the bat coronaviruses bind ACE2 compared to SARS-CoV-2. Using binding modeling and structural analyses, the researchers showed exactly how the bat virus receptor binding domain could interact with the ACE2 protein, offering a biochemical explanation of exactly how the bat Spike bind. The similarities between SARS-CoV-2 and the two bat viruses extend across the entire genome with one marked exception, Orf8. Within SARS-CoV-2 variants, Orf8 is among the most frequently mutated genes.
To understand the interaction of bat receptor-binding domains with ACE2 and the functional dynamics of the relationship, the researchers performed a biolayer interferometry assay with the different viruses to analyze the nanomolar range of binding affinity. In layperson terms, they tested to see if the bat viruses bound to ACE2 in a similar tightness to SARS-CoV-2, which they did.
Finally, to analyze the binding structure, they used x-ray crystallography at 2.9-angstrom resolution for both SARS-CoV-2 and BANAL-20-52. The figure below displays the structural similarities and differences between the two complexes, particularly the mutated residues in the receptor-binding domain and the structural complexes in the helices. As expected, most of the interactions observed between the ACE2 and SARS-CoV-2 receptor-binding domain were also present for BANAL-236.
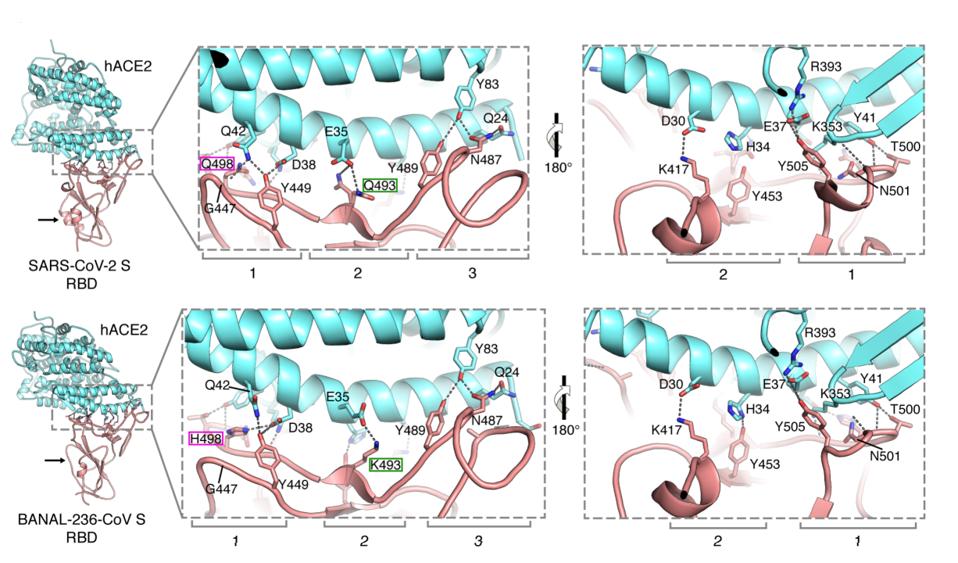
FIGURE 2: Ribbon representations of the crystal structures of hACE2 peptidase domain (cyan) in complex with SARS-CoV-2 (PDB 6M0J) or BANAL-236 (this study, PDB 7PKI) RBDs (pink). Black arrows in the overall structures indicate the structural
TEMMAM ET AL
Recombination is frequently observed among naturally occurring coronaviruses. The Laotian bat viruses are no exception. The authors were able to decipher recombination events amongst the broad family of coronaviruses infecting bats, which likely contribute to the origin of SARS-CoV.
The picture that emerges is a continual reassortment of genes leading to a mosaic of viruses. For both SARS-CoV-2 and the bat viruses, there appear to be two separate origins of the exterior Spike protein, one for the N-terminal domain and another for the receptor-binding domain.
The one question that remains is the origin of the Furin cleavage site. A recent survey of the viruses found that there are many coronaviruses with independently-derived Furin cleavage sites. Many genera of coronavirus, including alphacoronavirus, gammacoronavirus, and betacoronavirus contain some viruses with the naturally-derived site. We note that some viruses that pairs of viruses that are otherwise identical over their entire lengths may differ in the presence or absence of a Furin cleavage site. For instance, SARS-CoV-2 contains one, whereas SARS-CoV lacks one. In fact, the Furin cleavage site is often lost merely by growing SARS-CoV-2 in human tissue culture. It is likely that the Furin site arises in some SARS-CoV-2 variants and bat viruses by natural recombination, as it may have been the case for SARS-CoV-2.
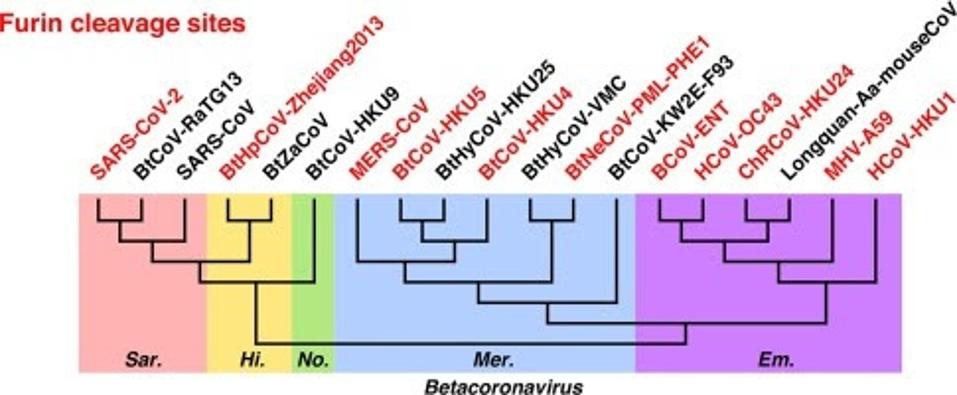
FIGURE 3: Furin cleavage sites at spike S1/S2 naturally occurred independently for multiple times in coronaviruses. Figure and legend from Yiran et al.
YIRAN ET AL
The authors speculate that the Furin cleavage site could be acquired upon coinfection with either another bat or some other exotic species. I would add here that it could have been acquired by coinfection with humans in the region previously coinfected with another bat virus which contains a Furin cleavage site in a separate recombination event,
What is the significance of this finding? As we have already experienced, the Covid-19 pandemic is not a one-off event. Humans are repeatedly infected and perhaps quite frequently infected with coronaviruses of bat and other species origin. It is believed that MERS-CoV, for example, was acquired from camels, which in turn was acquired from viruses prevalent in humans and bats in Africa. Many of the cold viruses that sweep through our population also have an animal origin, such as OC43, which originated in domestic farm animals like cows and pigs. In the recent past, there have been reports of canine coronaviruses infecting children in Indonesia. Moreover, SARS-CoV-2 variants have shown that coronaviruses can continually adapt to their host, resulting in a more infectious and pathogenic virus.
With respect to the current pandemic, we should be alert to the possibility of new, more transmissible, dangerous versions of COVID-19 may emerge. Many hundreds of millions have already been infected with SARS-CoV-2, as well as domestic and feral fauna. It is possible that new adaptations via animal coinfection may emerge, such as those found in mink. Additionally, recombination events between SARS-CoV-2 and other coronaviruses that exist in the human population could give rise to new and more dangerous forms of the pandemic.

