The Omicron Surprise
(Posted on Wednesday, February 2, 2022)
This is the third in a series on the Omicron variants.
The rise of Omicron has come as a surprise. Many were hoping Delta would be the last in a chain of variants of increasing transmission and virulence. Not one but two new forms of Omicron, BA.1 and BA.2, have put a swift end to that hope. Both seem to have arrived as if from nowhere to quickly sweep the world.
Omicron first appeared in Southern Africa in late November. Omicron appeared not as a single variant but as three simultaneously denoted BA.1, BA.2, and BA.3. All three are derived from a common parent, which itself derived from the long-vanished B.1 variant, which was the dominant global virus beginning in early 2020 until it was displaced by an alphabet of regional variants: the Alpha to Eta set.
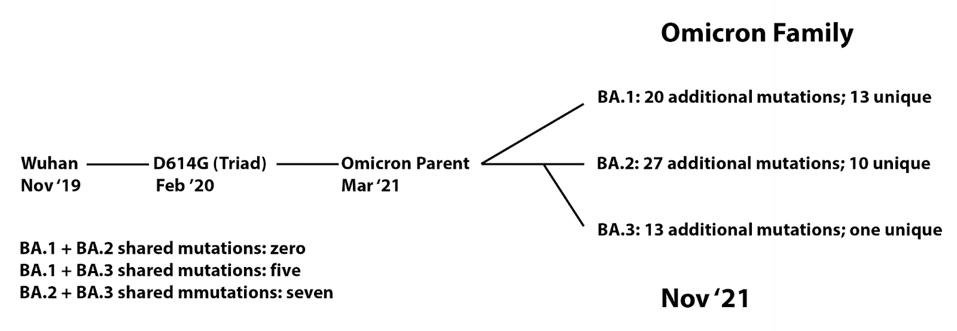
FIGURE 1: Lineage of Omicron family viruses. The date included is when that specific variant was first collected.
ACCESS HEALTH INTERNATIONAL
Despite detection simultaneously, all three Omicron variants are as different from one another as Alpha is from Beta, Beta from Gamma, and Gamma from Delta.
The origin is shrouded in mystery. Did all three incubate in an isolated population? Is the parent the result reverse Zoonosis? Did all three arise in one or more immunosuppressed individuals after prolonged incubation? Is there another story to be told?
Omicron Dominates
As I write in early February 2022, Omicron is now the dominant virus worldwide. BA.1 quickly displaced Delta in mid-rampage in many parts of the world, including the United States and Europe. Concurrently, BA.2 is on the rise or the dominant variant in many countries. When BA.1 and BA.2 are present, BA.2 is out-competing even BA.1. As of now, BA.3 accounts for only a few infections.

FIGURE 2: Regional distribution of cases relative to the frequency of Delta, BA.1, and BA.2 strains.
STEBBINGS
Sadly, most countries seem entirely helpless to stop the sweep of the virus. Omicron infections rise and fall not due to our efforts to contain the infection but rather from its own internal dynamics. To state it clearly, Omicron infections are out of our control (with the exception of China, as per usual).
The degree of transmissibility of both BA.1 and BA.2 is astonishing, outpacing even our most pessimistic projections by epidemiologists and virologists. Omicron leads to near-vertical increases in cases numbers. In the two months since Omicron was first discovered, the virus has infected at least 90 million people worldwide, more than all other variants for 2020, the first full year of the pandemic.
Several key novel characteristics seem to account for Omicron’s spread. The virus’s ability to evade immune protection conferred by prior infection or vaccination is startling. Both BA.1 and BA.2 rip through even well-vaccinated populations as if they were Covid naive. Once the infection is initiated, the virus takes hold at a record pace. Virus concentrations reach a peak within two days of exposure, assuring those who contract the virus spread it quickly to others.
Transmission
Just how very transmissible are the Omicron variants? The previously dominant variant Delta is about 1.5 times as transmissible as Alpha, Beta, and Gamma., In their day, the prior variants of concern were about 1.5 more transmissible than the original B.1 variant, which was somewhat more transmissible than the original Wuhan isomers. Recent studies from Denmark where BA.2 rapidly displaces BA.1 as the dominant variant suggest that BA.2 is at least 1.5 times more transmissible than BA.2. After some quick calculations, BA.2 is at least seven and a half times more transmissible than the original Wuhan isolate, on par with the most infectious diseases ever described!
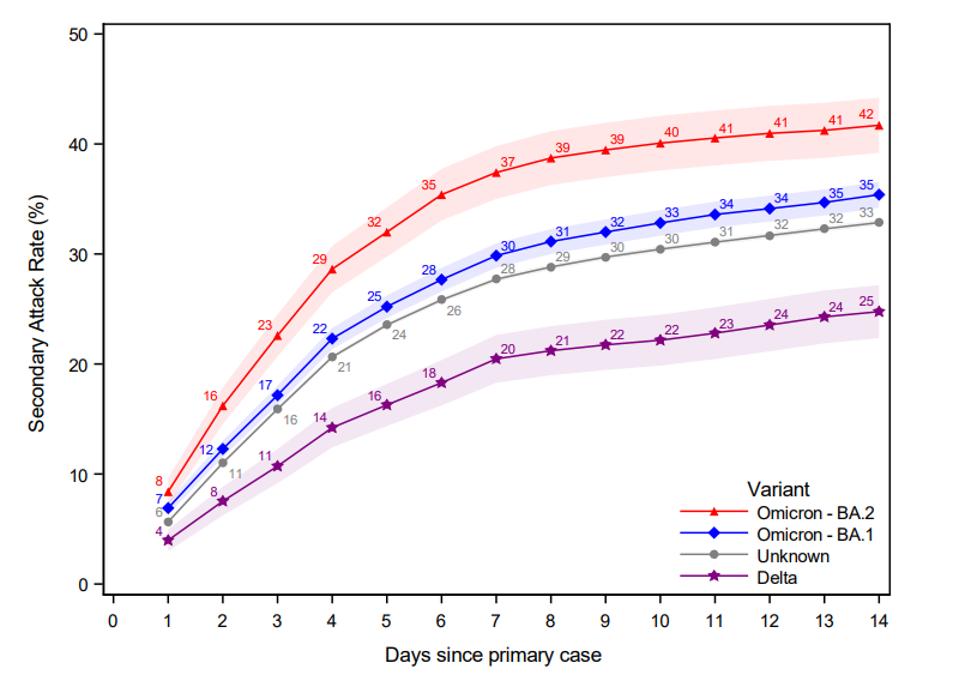
FIGURE 3: Secondary Attack Rate for households infected with the Omicron BA.1, BA.2, and Delta VOC, as well as those without an identified variant. The secondary attack rate was defined as the proportion of potential secondary cases within the same
LYNGSE ET AL.
We are fortunate that increased lethality does not track with Omicron increased infectivity. BA.1, though pathogenic, is less virulent than Delta. BA.1 infection of susceptible hamsters and mice causes a much milder disease than with earlier variants. How much less virulent Omicron BA.1 is than another variant—as measured by severe disease incidence, hospitalization, and death—is up for debate. What is clear is that full vaccination (three doses) or prior infection plus vaccination (two doses) reduces serious consequences of Omicron infection five to tenfold. A recent preliminary study from South Africa suggests that the unvaccinated Omicron is still about as virulent as the early Alpha variant. Be that as it may, the BA.1 variant, as of early February 2022, kills between 2,000 and 3,000 people a day in the United States alone. The bright note is that complete vaccination (three or four shots of an mRNA or a combination of an adenovirus-based vaccine with two doses of mRNA vaccines) provides substantial protection against hospitalization and death.

FIGURE 4: Severity of Disease in the Unvaccinated
ACCESS HEALTH INTERNATIONAL
Immune escape
The vast number of mutations in the Spike protein BA.1 and BA.2 (more than thirty each) alter the immune response to the virus. Sera of susceptible mice infected with BA.1 neutralize BA.1 effectively, but the same neutralize all prior variants poorly. Conversely, most sera induced either by natural infection or vaccination are approximately ten times less effective in neutralizing BA.1 than their activity against other variants. Current vaccines’ protection wanes to close to zero over five to six months.
Virus Entry
Efficient cleavage of the Spike protein while budding from the surface of a virus-producing cell is a hallmark of SARS-CoV-2 compared to SARS-CoV-1. Efficient cleavage of the precursor Spike proteins into the active S1 and S2 proteins allows SARS-CoV-2 to enter the cell via fusion of the viral cell membranes. In contrast, the endosomal entry route is typical of SARS CoV-1. Efficient cleavage of the SARS-2 spike is attributed to the furin cleavage site at the S1/S2 boundary. Spike protein cleavage at the cell also underlies syncytial formation (cell-to-cell fusion) of infected cells.
A puzzle: BA.1 retains the furin site but is poorly cleaved. Why? The answer must lie in the altered structure, most probably mutations near the cleavage site and in the membrane-associated S2 protein. Still, despite elegant work on the structure of the Omicron spike, the details remain a mystery.
Analysis of Omicron BA.1 and BA.2 Mutations
Orf1ab: Replication Complex
The replication complex of SARS-CoV-2 is comprised of 16 nonstructural proteins (NSP 1-16). Between BA.1 and BA.2, there are 21 changes in the amino acids in this otherwise highly conserved region of SARS-CoV-2. Of these seven are shared between the two, six are unique to BA.1, and eight are unique to BA.2 One of these shared mutations is a proline to leucine change at position 323 of the RNA-dependent RNA Polymerase (NSP12 P323L). This NSP12 mutation, together with the nucleotide change C241U in the five prime untranslated regions, is common to all variants of the B.1 lineage, including the Alpha, Beta, Gamma, and Delta. The functional significance of the single amino acid changes in the polymerase proteins is yet to be determined. The single nucleotide change in 5 prime translated regions is reported to create a binding site for the cellular TAR binding protein known to regulate RNA metabolism and increase the rate of virus replication.
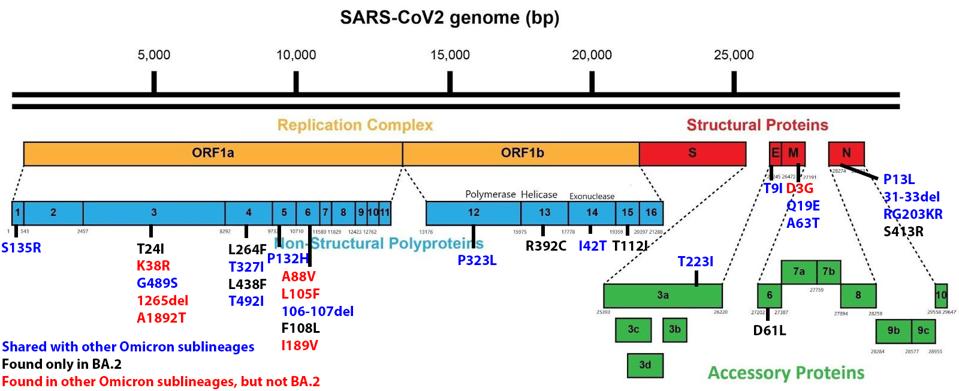
FIGURE 5: BA.2 Genome mutations. Those in blue are shared with other Omicron sublineages. Those in black are found only in BA.2.
ACCESS HEALTH INTERNATIONAL
Spike Protein
The Spike protein of the Omicron variants is heavily mutated. There are 43 amino acid changes between the sequence of the reference spike protein and that of the BA.1 and BA.2 proteins, far more than any other variants of concern. Of these, 22 are shared, 16 are unique to BA.1, and 5 are unique to BA.2.
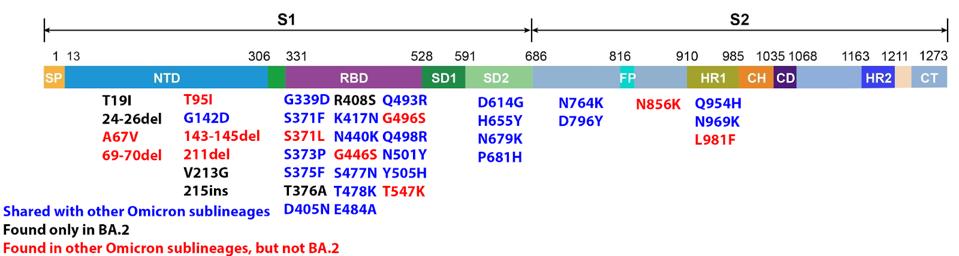
FIGURE 6: BA.2 Spike mutations. Those in blue are shared with other Omicron sublineages. Those in black are found only in BA.2. Those in red are found in other Omicron sublineages, but not BA.2.
ACCESS HEALTH INTERNATIONAL
Neutralizing antibodies in sera of infected patients recognize the receptor-binding domain (amino acids 331 to 528) and the N terminal domain (amino acids 13-306). Within the N-terminal domain of BA.1 and BA.2, there are 14 amino acid changes. Only one is shared between the two variants. Twelve are unique to BA.1, and three are unique to BA.2. Such divergence suggests that the two variants evolved resistance under different immune pressure, possibly in different hosts. Antibodies that neutralize the virus via binding N-terminal domain epitopes of BA.1 are unlikely to neutralize BA.2 and vice versa.
There are 18 amino acid changes in the receptor-binding domain of the two Omicron variants. Thirteen of these are shared. Two are unique to BA.1, and 3 are unique to BA.2. Likely, most antibodies that bind BA.1 will also bind and neutralize BA.2. I suspect most sera will be cross-neutralizing, whereas many monoclonal antibodies may neutralize only one but not both variants.
There are 11 amino acid changes in the Spike protein that lies carboxy-terminal to the receptor-binding domain. Eight are shared, and three are unique to BA.1. I suspect that BA.2 is likely to share the unusual cleavage and membrane fusion properties of BA.1.
The Structural Proteins M, E, and N
All genes that encode the structural proteins S (spike), M (membrane), E (envelope), and N (nucleocapsid) are located three prime to orf1ab. BA.1 and BA.2 share the same single amino acid mutation in the envelope protein. Two of the three amino acid changes in membrane protein BA.1 contains one additional amino acid change in the membrane protein.
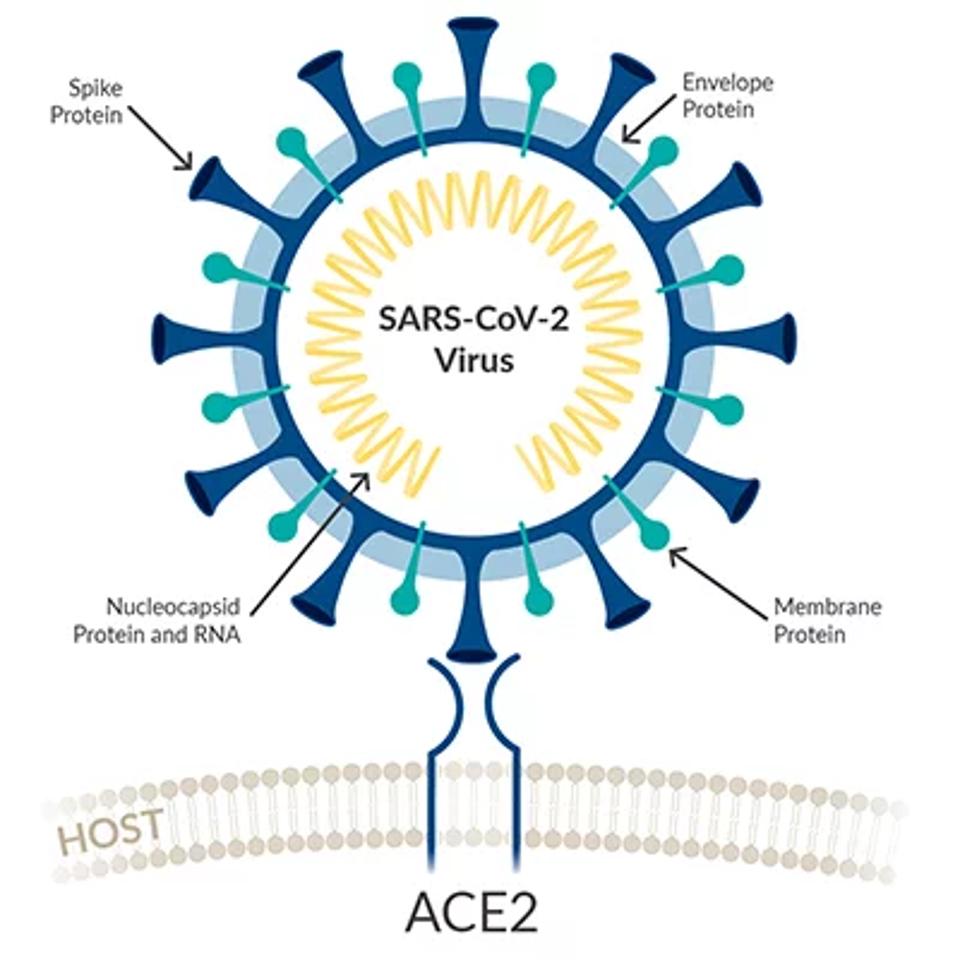
FIGURE 7: Viral schematic of SARS-CoV-2 structural proteins.
ACCESS HEALTH INTERNATIONAL
These changes entail either a charge or a change from polar to non-polar residues, potentially affecting the structure and function of the mature virus particle. These changes may increase the stability of the virus particle and therefore prolong the survival of the virus in the air or on surfaces. Increased stability of the virus particle may also increase the transmission rate.
There are seven amino acids changes in the nucleocapsid proteins. Six of these are shared between BA.1 and BA.2. One is unique to BA.2. Two of the changes are common to most variants of concern, those at amino acids 203 and 204. Identical changes are found in most variants of concern. These changes have been shown to dramatically increase the titers of virus when green on cells in the laboratory and may account in part for the increase in transmission of all variants as compared to the reference isolates. BA.1 and BA.2 share the same deletion of amino acids 31, 32, and 33. This deletion is unique to Omicron.
Accessory Genes
SARS-CoV-2 specifies an additional set of accessory genes, many of which regulate the host’s innate immune response. These include Orf3a,b,c and d; Orf6, Orf7a and b; Orf8; Orf9b; and Orf10. There are no amino acid changes in these proteins in BA.1 relative to the reference isolate and only two in BA.1 proteins—a single amino acid change in Orf3a and another in Orf6 of BA.2. The lack of mutations in the accessory proteins is usual amnesty variants of interest.
Noncoding Mutations
We also note many noncoding mutations in the Omicron viral genome. These nucleotide changes that do not change amino acids could still impact the virus. There are sequences throughout the virus that play into processes like replication and transcription. Noncoding mutations have the potential to impact these sequences. Below is the summary of noncoding mutations in BA.2.

FIGURE 8: Noncoding Mutations in Omicron sublineages.
ACCESS HEALTH INTERNATIONAL
Ultimately, BA.2 poses a similar, if not more significant, threat than its more pervasive sibling BA.1. All indications suggest at least 1.5 times as transmissible as BA.1. If the virulence of this strain remains the same or is more severe, it could yield more infections, hospitalizations, and deaths than any previous point of the pandemic. As such, we must remain cautious and stringent in our control of Covid-19.
The lesson from Omicron is that we should expect the unexpected from this wily virus. We are just beginning to take its full measure. What will come next, a variant that has the lethality of SARS (killing 4% of the young and 45% of the elderly) that transmits like Omicron, or a variant that spreads quickly, evades immunity but causes only cold-like symptoms? Regardless of what is to come, we must do better at controlling the pandemic by all measures possible, public health (detection, contact tracing, and isolation for the incubation period), near-universal vaccination, development of prophylactic and therapeutic drugs, and global equity in access to vaccines, diagnostic tests, and vaccines.

