Progress In The Search For Broadly Neutralizing Monoclonal Antibodies V
(Posted on Monday, August 1, 2022)
This is part of a continuing series describing antiviral antibodies to prevent and treat SARS-CoV-2 infections. In this series, we will discuss the fundamental nature of virus evolution, how SARS-CoV-2 has mutated to evade neutralizing antibodies, and our latest attempts to fight against these mutations with more recent and improved antibody candidates.
As novel SARS-CoV-2 variants develop new mutations, their evasion from existing treatments and vaccines continues to increase. Antibodies from vaccines wane after a few months, antibodies from previous infections are often ineffective, and monoclonal therapies that worked against earlier variants struggle against current strains. The current state of the pandemic has sparked a search for monoclonal antibodies that neutralize not one but all strains to counter these new variants.
In this series, we have discussed several pan-variant monoclonal antibodies, all of which promise against current Omicron strains and previous variants of concern such as Alpha, Beta, and Delta. Here we analyze another described in a study by Dacon et al.: the COV44-62 and COV44-79 antibodies.
COV44-62 and COV44-79 Antibody Origin
Researchers from the National Institute of Health and Scripps Research Institute began their search for monoclonal antibodies by a traditional method: examining the plasma samples of 142 previously infected donors. They scanned these samples not only for SARS-CoV-2 recognition but also for six other human coronaviruses, including SARS-1 and MERS. Of the 142 samples, 19 recognized SARS-CoV-2 and at least two other betacoronaviruses. By finding antibodies that recognize multiple betacoronaviruses, the likelihood increases of finding antibodies that recognize various strains of SARS-CoV-2 as well.
From these 19 samples, they identified a staggering 673,671 IgG B cells. They tested for reactivity and binding specificity to a panel of coronavirus Spike proteins to narrow this pool. They eventually found a set of six monoclonal antibodies that bound all seven human coronaviruses tested.
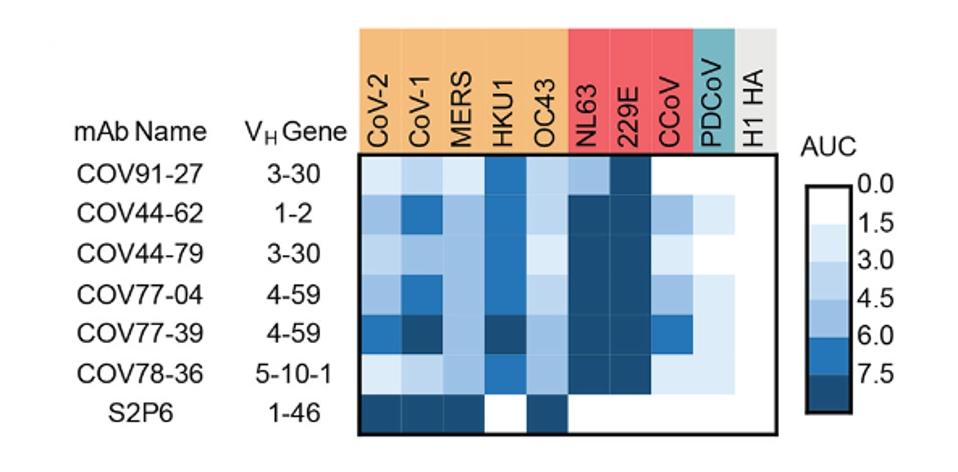
FIGURE 1: Heat map representing the binding of broadly reactive mAbs to spike proteins from coronaviruses across the alpha, beta and deltacoronavirus genera.
DACON ET AL.
COV44-62 and COV44-79 Antibody Neutralization
Introducing the set of six monoclonal antibodies to a neutralization assay of pseudotyped betacoronaviruses, Dacon et al. found that two antibodies, COV44-62 and COV44-79, showed the broadest functional neutralization, disabling SARS-CoV-2, SARS-CoV-1, and HCoV-OC43, as well as the alphacoronavirus HCoV-NL63 and HCoV-229E.
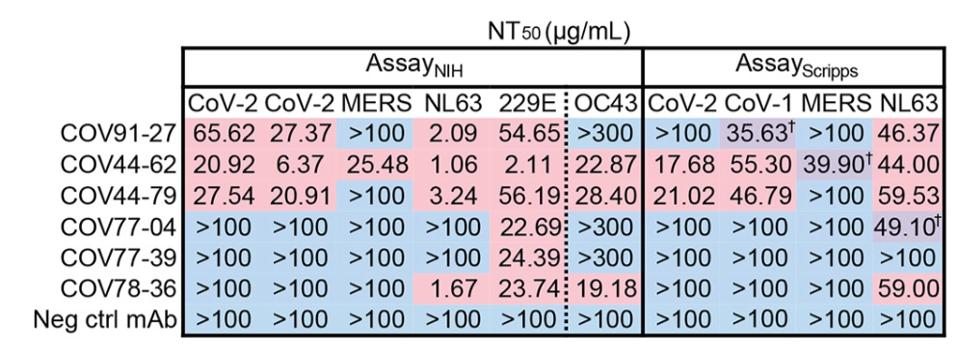
FIGURE 2: Antibody titer neutralization of pseudotyped coronaviruses.
DACON ET AL.
Perhaps more notably for our search, both COV44-62 and COV44-79 strongly neutralized a host of SARS-CoV-2 variants. In more pseudotype assay neutralization tests, COV44-62 and COV44-79 effectively neutralized Alpha, Beta, Gamma, Delta, Mu, Omicron BA.1, BA.2, and BA.4/5. Neutralization of Omicron BA.4/5 is significant as they are still the majority sequence fueling infections worldwide as this is written in late July. In general, COV44-62 achieved neutralization at lower concentrations, indicating a more efficient antibody, but we note that COV44-79 neutralizes BA.4/5 more efficiently than COV44-62.
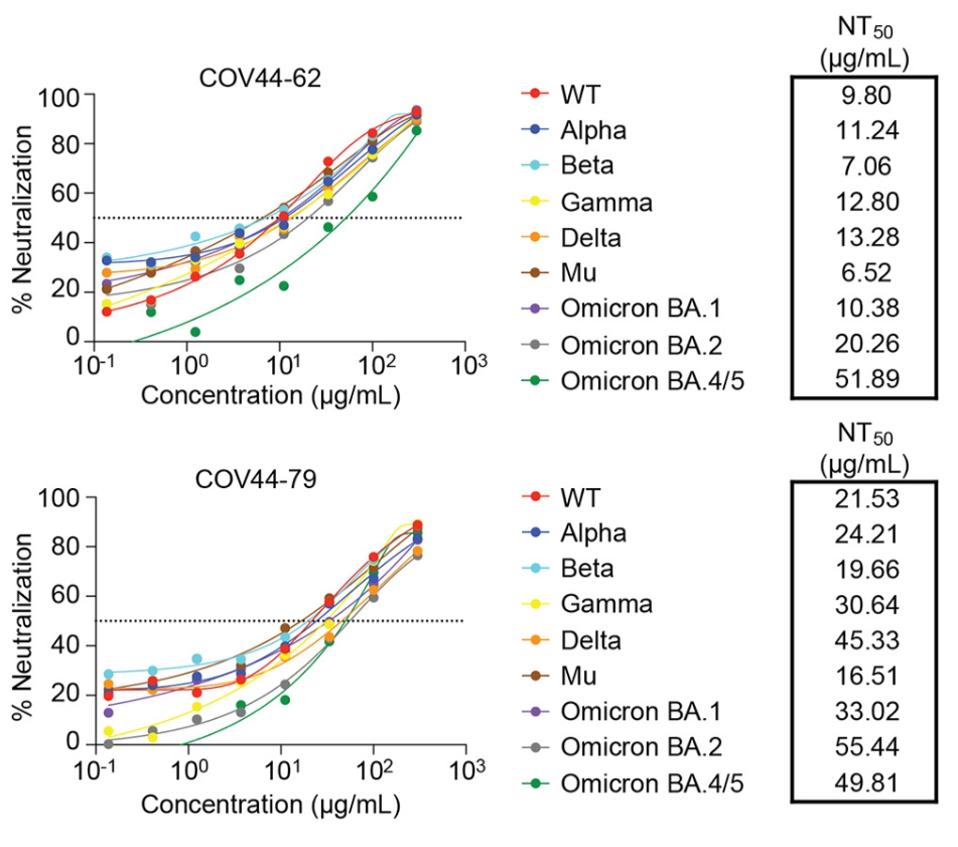
FIGURE 3: Neutralization of SARS-CoV-2 variants of concern (pseudovirus) by COV44-62 and COV44-79.
DACON ET AL.
COV44-62 and COV44-79 Antibody Target
While the rest of the antibodies in this series have also neutralized many variants of concern, what makes these two antibodies particularly special is their variant neutralization and Spike residue target. Most monoclonal antibodies target amino acids on either the receptor-binding domain, the N-terminal domain, or a combination of the two. These two domains facilitate ACE2 binding to the host cell, and blocking these connections is often an effective way to prevent host cell infection.
The COV44-62 and COV44-79, as well as the other four antibodies from the broadly recognizing set, all prefer to bind in the S2 portion of the Spike, specifically the fusion peptide. The two antibodies bind from amino acid positions 812-830 in the SARS-CoV-2 Spike protein.
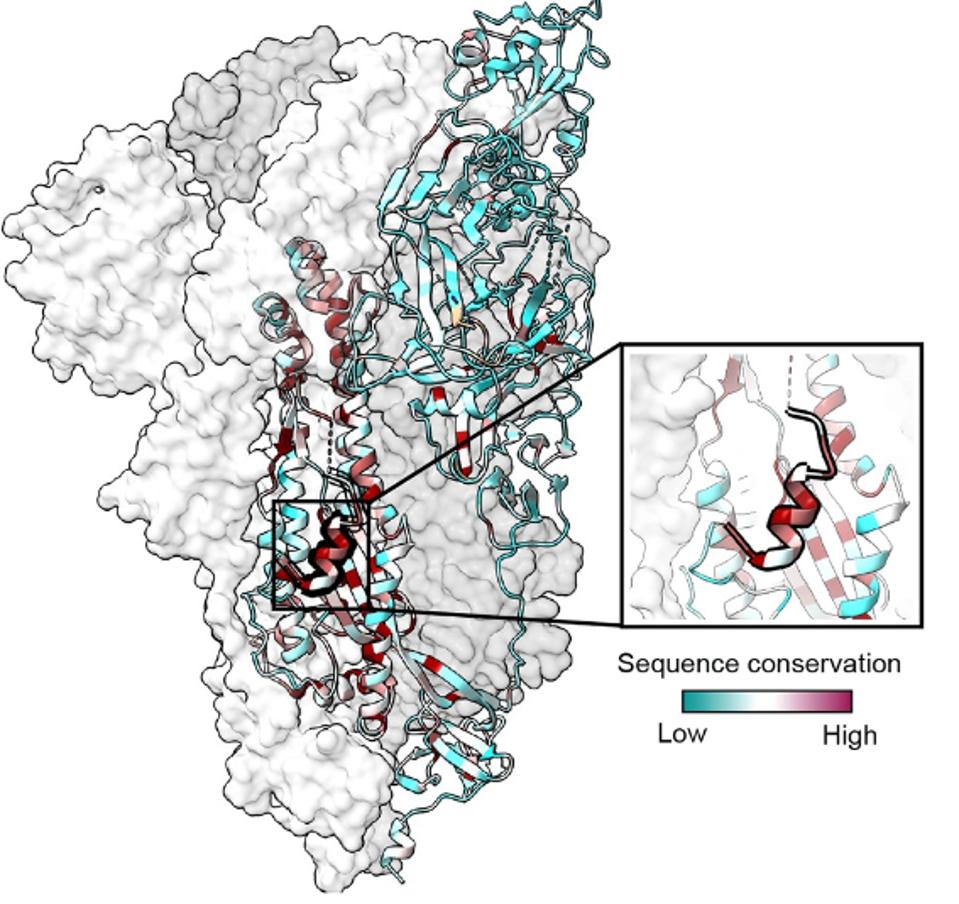
FIGURE 4: Fusion peptide conserved sequence in the SARS-CoV-2 Spike protein.
DACON ET AL.
Dacon et al. note that critical binding residues include R815, E819, D820, L822, and F823. These five residues are amongst the most conserved in the coronavirus genera Spike protein, all of which are conserved in at least 34 of 35 coronavirus species. This also holds true for SARS-CoV-2 variants of concern; none carry mutations at these five amino acids. In fact, in the GISAID SARS-CoV-2 sequence database, all five mutations are identified less than 5,000 times among 12 million sequences, many of which are likely dead viruses.
Early in vivo trials on Syrian hamster models found that those treated with COV44-79 recovered from moderate to severe symptoms within 3-7 days, and those treated with COV44-62 recovered in 5-7 days.
As we recommend with all broadly neutralizing antibodies, there is no reason to limit treatment to just one. An antibody cocktail of two or three monoclonal antibodies covering a broad footprint of conserved residues could be a powerful weapon against current and future strains, which are sure to continue mutating to evade immunity. We must prioritize and expedite these antibodies’ production as the pandemic continues to rage.

