Progress In The Search For Broadly Neutralizing Monoclonal Antibodies VI
(Posted on Thursday, September 1, 2022)
This is part of a continuing series describing antiviral antibodies to prevent and treat SARS-CoV-2 infections. In this series, we will discuss the fundamental nature of virus evolution, how SARS-CoV-2 has mutated to evade neutralizing antibodies, and our latest attempts to fight against these mutations with more recent and improved antibody candidates.
SARS-CoV-2 variants that evade the immune system are characteristic of the Covid-19 pandemic. As novel SARS-CoV-2 variants develop new mutations, their evasion from existing treatments and vaccines continues to increase. Antibodies from vaccines wane after a few months, antibodies from previous infections are often ineffective, and monoclonal antibody therapies that worked against earlier variants struggle against current variants. The current state of the pandemic has sparked a search for monoclonal antibodies that neutralize not one but all variants to counter these new variants.
In this series, we have discussed several pan-variant monoclonal antibodies, all of which promise against current Omicron variants and previous variants of concern, such as Alpha, Beta, and Delta. Here we analyze two new antibodies described in a study by Low et al. that target a conserved site not only in SARS-CoV-2 variants, but in many similar coronaviruses as well.
Antibody Discovery
Low et al. began their search in a familiar fashion: isolating memory B cells from convalescent and vaccinated SARS-CoV-2 patients. This yielded many thousand samples from 43 individuals.
Next, the researchers introduced the samples to an ELISA panel of many viruses, testing for cross-reactivity to a broad range of coronaviruses. The tested viruses included SARS-CoV, MERS-CoV, human coronaviruses OC43, HKU1, NL63, and 229E. A group of seven antibodies emerged cross-reacted against all Spike proteins tested, including alphacoronaviruses NL63 and 229E. The antibodies were sorted into three groups depending on their broad binding capacity, with Group 3 designated for those that bound all tested coronaviruses
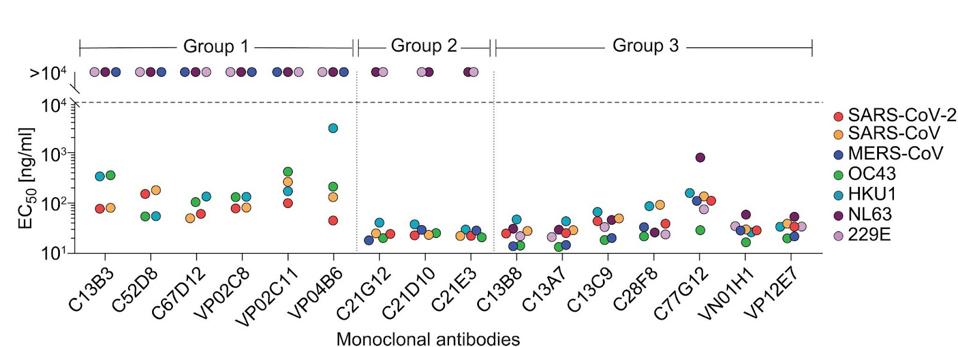
FIGURE 1: Monoclonal antibody cross-reactivity of a panel of coronaviruses. The colored dots represent coronavirus binding EC50 values, with the dashed line indicating the threshold for binding. Group 3 broadly cross-reacts with all viruses tested.
LOW ET AL.
Antibody Neutralization
The next step was to examine those that broadly bound for neutralizing capability. Testing all seven Group 3 antibodies for neutralization capabilities, Low and colleagues narrowed their search to two: C77G12 and VN01H1. The VN01H1 antibody neutralized all viruses examined, specifically SARS-CoV-2, SARS-CoV, MERS-CoV, NL63, and 229E from the binding assay and bat virus WIV-1 and merbecovirus PDF-2180. C77G12 neutralized the binding assay betacoronaviruses but to the highest degree of all Group 3 antibodies.
Hamster Study
Extending their studies to in vivo examination, they found that the Gamma variant SARS-CoV-2 virus in infected hamsters was potently neutralized by both antibodies, as were authentic Omicron BA.1 and BA.2 viruses. The researchers found that prophylactic administration of either C77G12 or VN01H1 reduced viral RNA copies and lung titers and ameliorated lung pathology at statistically significant levels.
Notably, earlier SARS-CoV-2 variants such as Alpha, Beta, and Delta were not examined in the study, nor were later Omicron variants, including the currently dominant BA.5 or BA.2.75 lineages.
Antibody Binding Site
Low et al. examined the exact binding of the antibodies by cryo-electron microscopy, a method of freezing small proteins and enzymes to enable a high-definition image of very small biological masses. Such broad cross-reactivity with a wide variety of SARS-CoV-2 relative viruses would require a highly conserved region of the Spike to bind. C77G12 and VN01H1 bind amino acids 811 through 825, which are highly conserved among all genera of the Orthocoronavirinae subfamily.
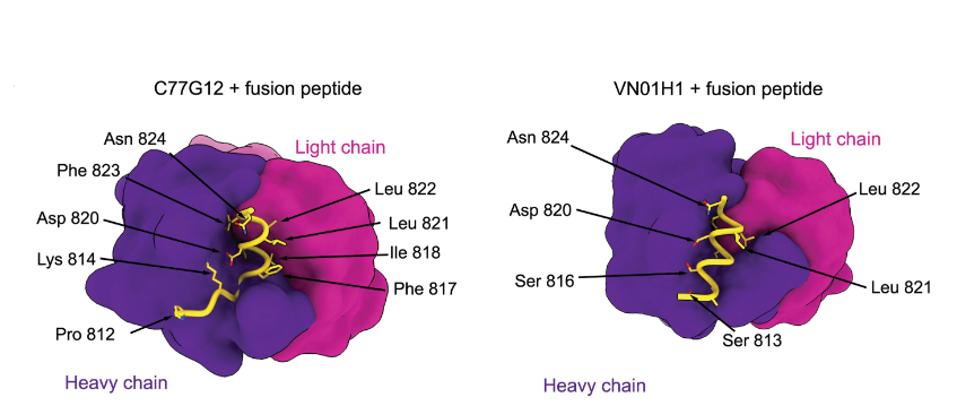
FIGURE 2: C77G12 and VN01H1 fusion peptide binding map.
LOW ET AL.
This conservation holds with all circulating SARS-CoV-2 variants to date. In the GISAID SARS-CoV-2 database containing 12.8 million sequences, the antibody binding epitope amino acids are only mutated a few thousand times.
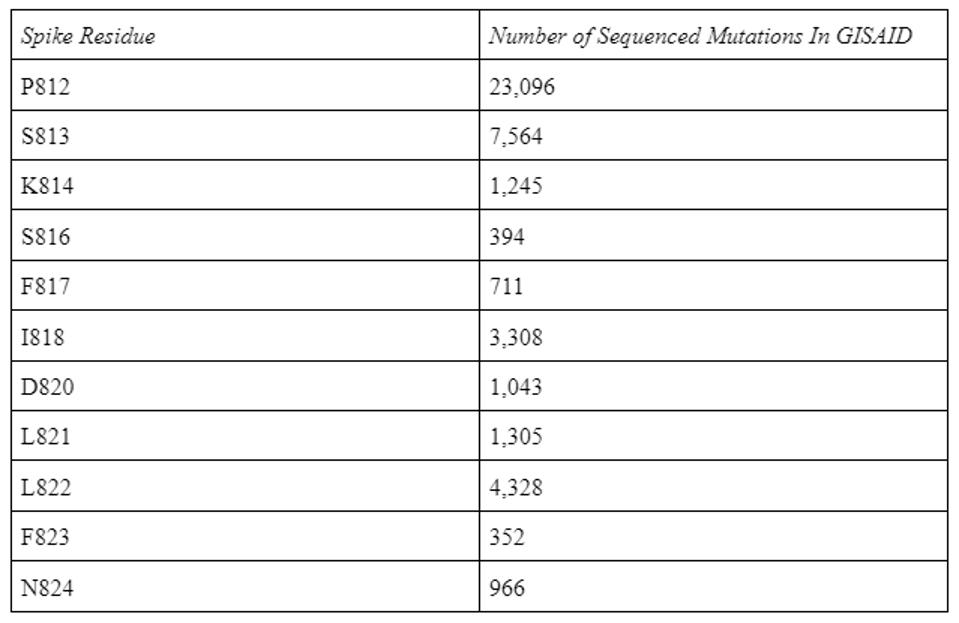
TABLE 1: Antibody epitope residue mutations in the GISAID database as of 8/31/2022.
ACCESS HEALTH INTERNATIONAL
The Low antibodies share a very similar epitope to the Dacon antibodies, described in a previous entry in this series. Both target the highly conserved fusion peptide and many of the amino acid contact points overlap between the two, and as such, both are viable candidates for monoclonal treatment in the months to come.
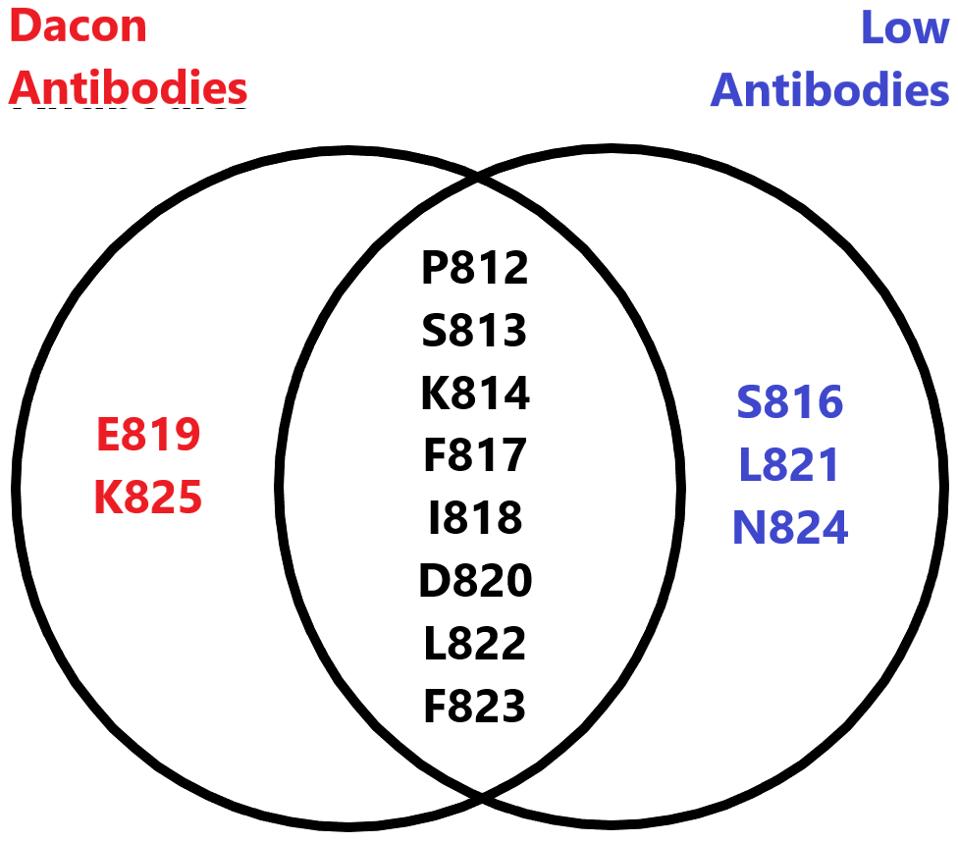
FIGURE 3: Venn diagram comparing the amino acid footprint of the Low antibodies and the Dacon antibodies.
ACCESS HEALTH INTERNATIONAL
Notably, the epitope target is buried toward the core of the Spike trimer and is highly inaccessible.
How, then, do these antibodies bind a hidden target? Low and colleagues identified a crack in the fusion peptide’s defenses using transfected human embryonic kidney cells. During ACE2 binding, conformational shifting of the Spike trimer exposes the cryptic epitope. The coronavirus Spike can be in one of two conformations based on its infecting host stage. The “down” conformation is before ACE2 binding occurs, and the “up” conformation is as ACE2 binding occurs. The receptor-binding domain attaches to the ACE2 receptor, revealing the fusion peptide, at which point C77G12 and VN01H1 can bind.
Antibody-Mediated Phagocytosis
Based on personal communications with the authors, we also note preliminary evidence that the antibodies may induce antibody-dependent cellular phagocytosis. ADCP is a mechanism of elimination by which the monoclonal antibodies target cells to promote clearance from the body by phagocytic immune cells. The phagocyte engages with the antibody and engulfs the infected cell, leading to quicker, more favorable outcomes for the patient.
Summary
The Low study adds another useful pair of broadly neutralizing monoclonal antibodies to our growing armamentarium of SARS-CoV-2 antibody treatments and prophylactics. In tandem with the Low antibody, additional antibodies could be used in combination to target other binding epitopes or trigger conformational changes to maximize antibody potency. Combination antibody therapies present the best weapon to overcome the virus and clear infected cells in Covid-19 patients. We recommend immediate high scale production of such combinations as the pandemic continues to rage in its third year.

