Will The BA.4.6 Variant Drive The Next Wave Of The Covid-19 Pandemic?
(Posted on Friday, September 16, 2022)
Here, we draw attention to a variant, BA.4.6, which is the latest variant spreading in the United States. We note in this summary that BA.4.6 contains five amino acid changes distinct from the currently dominant BA.5 variant and seventeen nucleotide changes, which all may act to improve viral fitness and trigger a new wave of the pandemic.
The World Health Organization sees the light at the end of the tunnel. The expression that comes to mind for Covid-19, stemming from the days of the Vietnam War protests, recalls a different perspective: I see a tunnel at the end of the tunnel. In particular, given our current state of understanding of viral variants, there is reason to believe that the next few years will closely resemble our recent experience of successive waves of new variants, each well adapted to spread in a previously vaccinated and/or infected population.
Coronaviruses are very well adapted to reinfection of adult immunocompetent mammals that have been previously and recently infected by their predecessor, such as the Influenza virus. In this respect, we should constantly look for new variants that may trigger yet another wave. Figure 1 summarizes the contemporary history of SARS-CoV-2 variants.
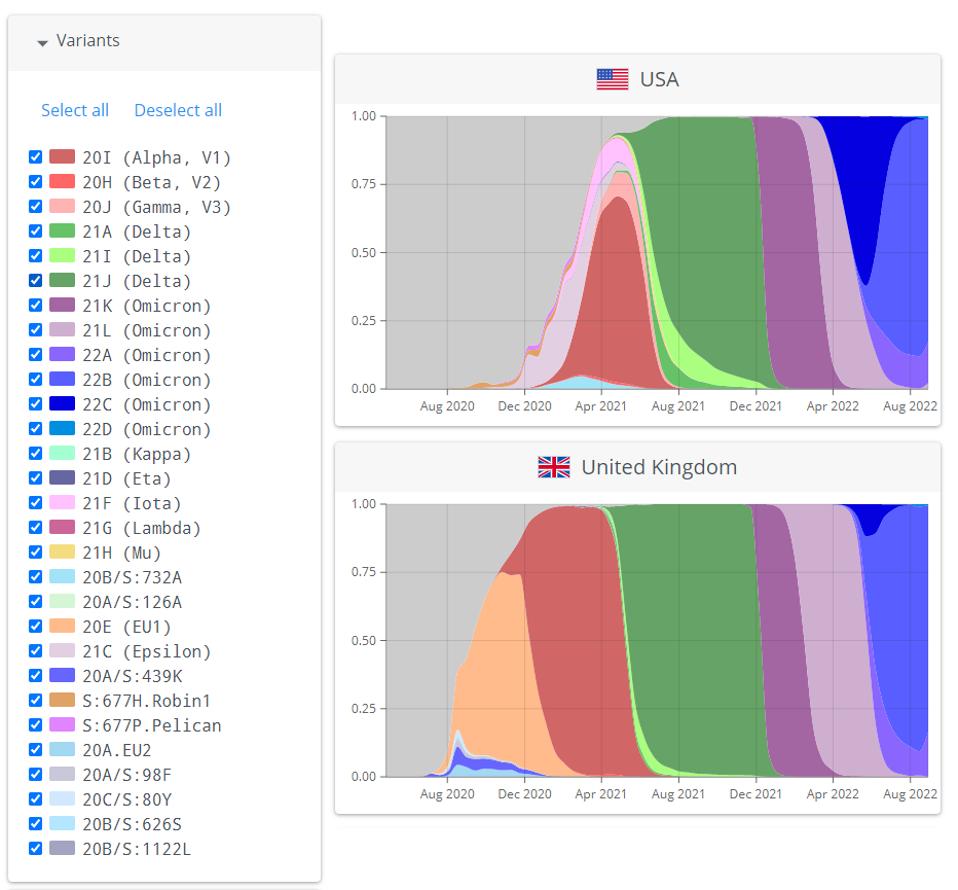
FIGURE 1: Successive waves of SARS-CoV-2 variants in the United States and the United Kingdom.
NEXTSTRAIN.ORG
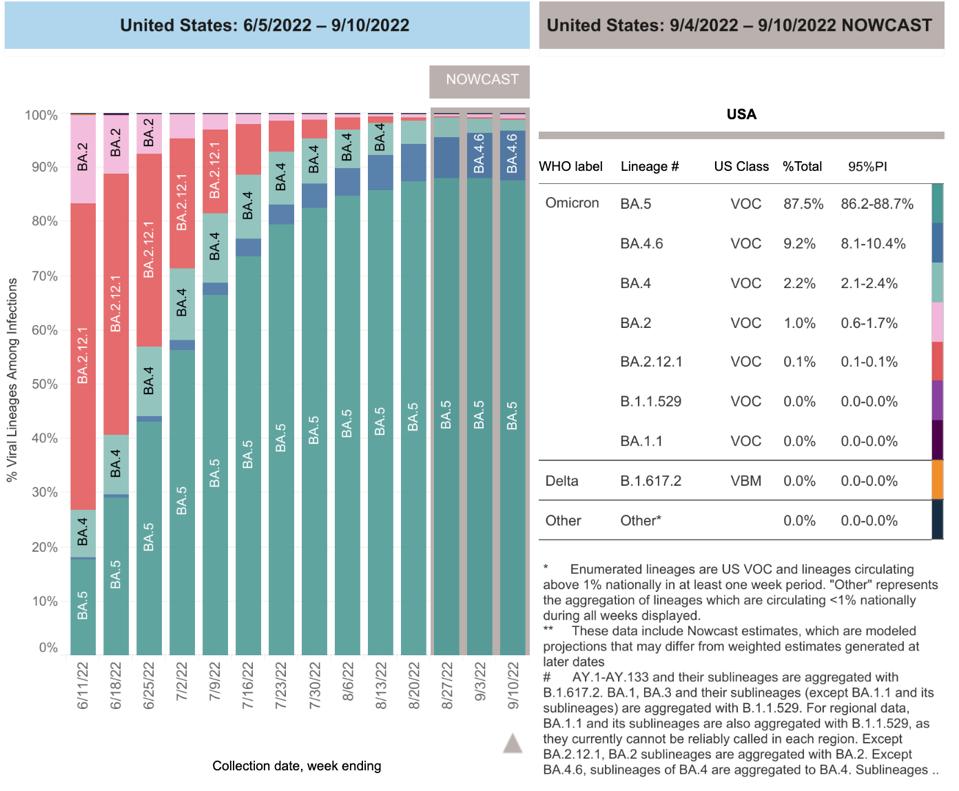
FIGURE 2: BA.4.6 gaining ground on BA.5 in the United States
CDC
BA.4.6 and BA.5
Between the currently rampant BA.5 and the emerging BA.4.6, there are only seven amino acid differences throughout the entire genome. Although the focus is typically on the spike protein, five of seven lie outside the spike. These include a deletion in NSP1, an amino acid in the membrane protein that is unmutated in BA.4.6 but is changed in BA.5, and three amino acid substitutions in Orf6, Orf7b, and the nucleocapsid (Figure 3).
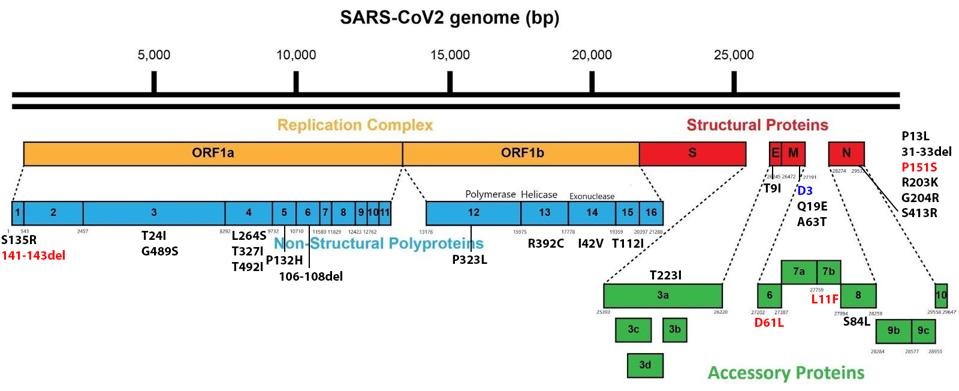
FIGURE 3: BA.4.6 Genomic mutations. Additional mutations in BA.4.6 as compared to BA.5 are highlighted in red. D3 in blue is unmutated, where it is mutated to D3G in Omicron BA.5.
ACCESS HEALTH INTERNATIONAL
In NSP1, BA.4.6 contains a deletion from positions 141-143. This mutation is observed in its parent, BA.4, but not in other Omicron variants. All Omicron variants contain the nearby mutation S135R. The deletion may affect NSP1’s immune suppressive functions, including mRNA translation efficiency, interferon inhibition, and phosphorylation inhibition of STAT1 and STAT2.
In the membrane protein, we note a significant difference between BA.5 and BA.4.6. In The prevalent BA.5, we note the D3G mutation, which is not found in BA.4.6. This difference may impact RIG-I/MDA-5 signaling, MAVS binding, and inhibition of downstream TBK1 signaling.
In Orf6, BA.4.6 contains D61L, which is common to all Omicron variants barring BA.5. While BA.5 is the most pervasive Omicron variant currently, that does not necessarily mean D61L in Orf6 detracts from the virus’s virological capabilities. This mutation may impact interferon inhibition, inhibition of nuclear translocation of STAT1, inhibition of TBK1 and IRF3, inhibition of nuclear translocation of IRF3, inhibition of karyopherin, and blockage of nuclear import and export.
In Orf7b, BA.4.6 contains L11F, which again comes from the parental BA.4. The mutation is absent in other Omicron variants. Orf7b is theorized to inhibit certain immune responses, but these are structural assumptions rather than experimental deductions, so further research is needed on this protein and mutation.
In the nucleocapsid protein, we observe P151S, which comes from BA.4 as well and is absent in other Omicron variants. The N protein is a significant player in inhibiting the host’s innate immune response. P151S may enhance interferon inhibition and RIG-I suppression of phosphorylation and nuclear translocation of STAT1 and STAT2.
Another mutation of note lies in the NSP5 protease. NSP5, or M Pro, has been shown in SARS-CoV-2 to be much more active in cellular protein cleavage than in previous coronaviruses. The mutation, P132H, is native to Omicron variants and may affect a critical region of the virus’s M Pro, potentially increasing the efficiency of cleavage of the viral proteins and regulatory proteins NEMO, optineurin, and RIPLET. These proteins are central to the activation of interferon-stimulated genes. The amino acid change imposes structural constraints on this critical region. The impacts of mutations in NSP5 are worthy of a more detailed study.
The BA.4.6 genome differs from BA.5 in the spike protein by only two amino acid mutations: R346T and N658S (Figure 4).
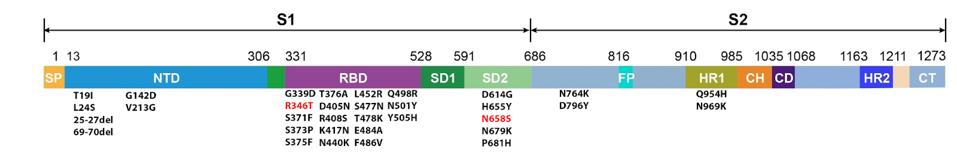
FIGURE 4: BA.4.6 spike protein mutations. Additional mutations in BA.4.6 as compared to BA.5 are highlighted in red.
ACCESS HEALTH INTERNATIONAL
Position 346 lies in the receptor-binding domain, the contact region between the virus and the host ACE2 receptor. Mutations in this region can impact transmissibility by increasing ACE2 affinity and may also boost immune evasion by blocking certain antibody binding sites on the spike.
Position 658 lies close to the S1/S2 cleavage point and may impact cleavage efficiency, though little is known about its exact impact.
Below is a Venn diagram demonstrating the overlap between BA.4.6 and earlier Omicron variants, namely BA.5 and BA.2.12.1 (Figure 5). Note the high degree of overlap between BA.4.6 and BA.2.12.1, as BA.4 is derived from the BA.2 lineage.
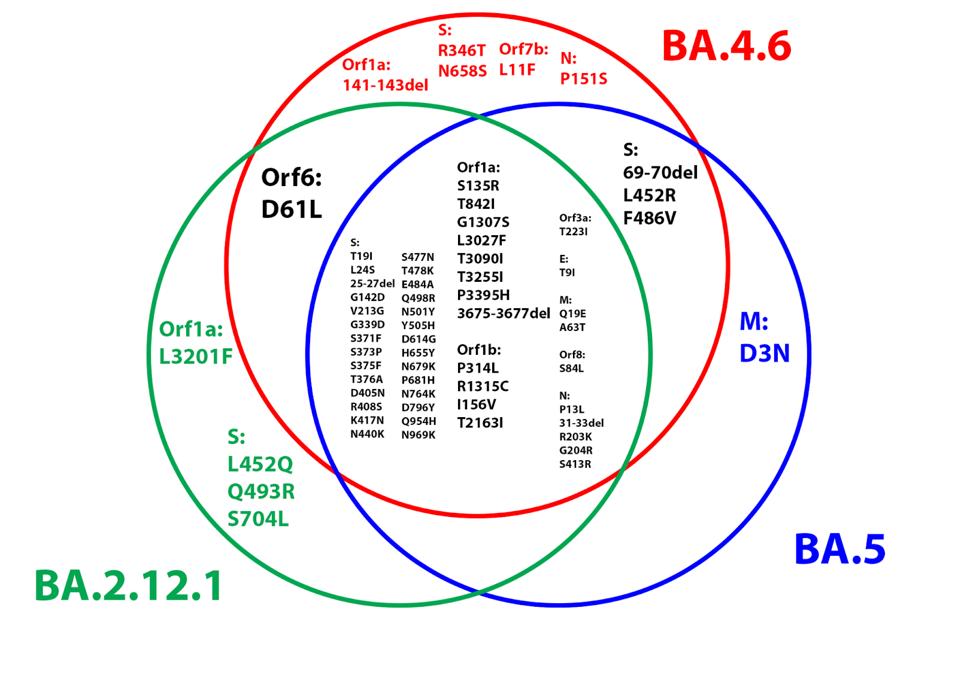
FIGURE 5: Venn diagram comparing the amino acid substitutions in the BA.4.6, BA.1, and BA.2.12.1 genomes.
ACCESS HEALTH INTERNATIONAL
We also note several synonymous mutations throughout the genome that do not impact amino acids but may still play an essential role in virulence and immune regulation via transcription sequences and other avenues (Figure 6). There are four synonymous mutations in NSP3, two in NSP12, and one in each of NSP2, NSP4, NSP5, NSP8, NSP10, NSP15, M, NSP6, NSP7b, and N. Research by Thorne et al. indicates that these mutations can initiate significant regulatory effects, particularly in structural and accessory proteins. These mutations are in addition to one mutation, C241T, present in all variants in the 5’ untranslated region. This section of the virus is difficult to analyze as it is not typically recorded.
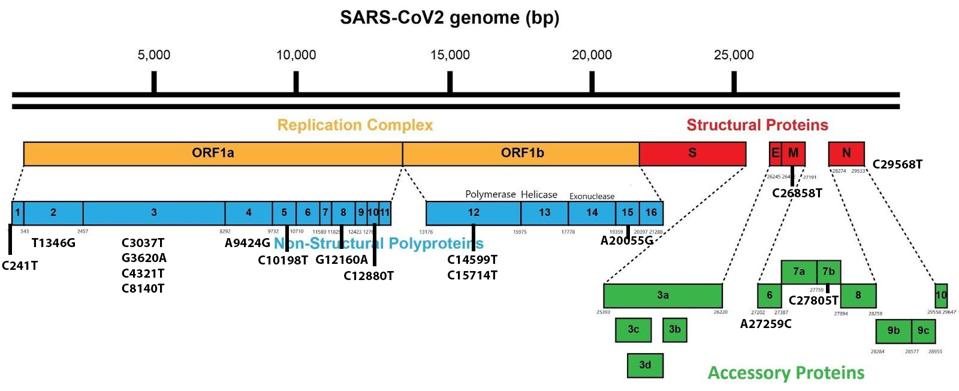
FIGURE 6: BA.4.6 Synonymous mutations.
ACCESS HEALTH INTERNATIONAL
We note that the ability of the virus to strengthen immune suppression may be directly related to the fitness of the virus in the population. Viral fitness is akin to a Rubik’s Cube. As the Cube requires each square to be in the correct formation to complete the puzzle, variants may have different mutations working epistatically to yield more efficient viral fitness.
BA.4.6 Immune Resistance
While BA.4.6 is still relatively uncommon in terms of global infections, some researchers have noted it as a variant worth examining, specifically in reference to its evasion of commercially available antibody treatments.
Jian et al. of Peking University in Beijing tested BA.4.6 against a panel of therapeutic monoclonal antibodies alongside its parent BA.4 (Figure 7). Of the 15 singular and combination treatments, only seven treatments displayed any degree of neutralization. Only three displayed substantial neutralization: Bebtelovimab, Cilgavimab, and the Tixagevimab + Cilgavimab combination.
Against BA.4.6, the results are even further diminished. Both Cilgavimab and the Tixagevimab + Cilgavimab combination lose all neutralizing capacity against BA.4.6. Six antibodies display at least minor neutralizing capacity, but only one, Bebtelovimab, strongly neutralizes the variant.
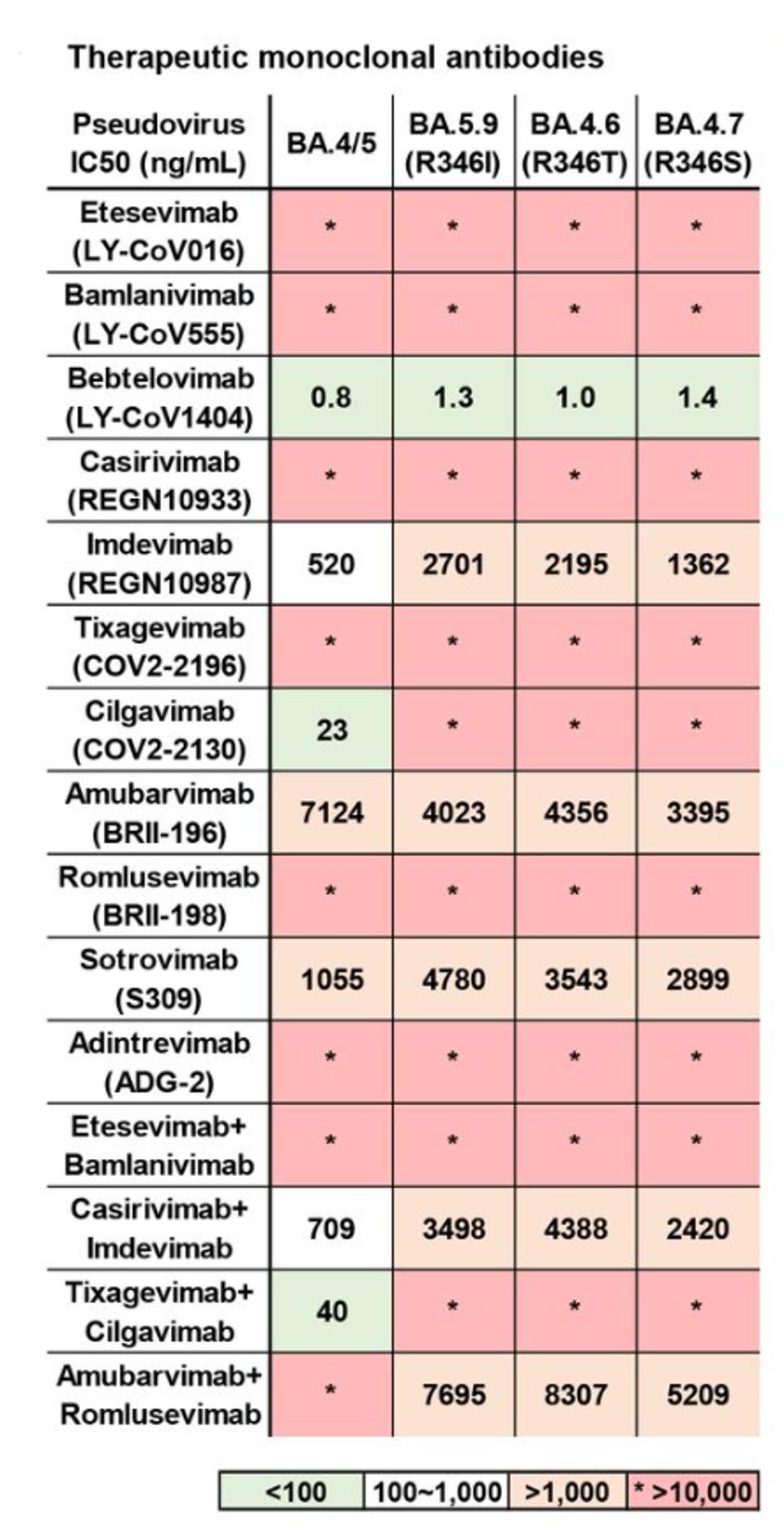
FIGURE 7: Neutralizing activities against SARS-CoV-2 D614G and BA.4/5 subvariants pseudovirus of therapeutic neutralizing antibodies. Background colors indicate neutralization levels. Green, IC50 < 100 ng/mL; white, 100 ng/mL < IC50 < 1,000
JIAN ET AL.
While we are lucky that we still have an available antibody that neutralizes the variant, continued use of Bebtelovimab may result in the mutation of variants to overcome the treatment, similar to weeds evolving to overcome specific weed killers. We must continue developing new treatments that maintain a wide variety of targets if we control the spread of emerging variants.
Broadly Neutralizing Monoclonal Antibodies
The next step for Covid-19 treatment will be the mass production of monoclonal antibodies, both singularly and in combination. Although the data on their ability to neutralize BA.4.6 is not yet available, it is noteworthy that the binding sites for many broadly neutralizing antibodies are conserved in BA.4.6, except for Luo et al., Fenwick et al., and Wang et al. We have previously pointed out the importance of using combinations. Specifically, we may use a variety of broadly neutralizing antibodies that minimizes mutations in amino acid binding sites. For example, the combination of Li et al., Dacon et al., and Bebtelovimab, which we know, neutralizes BA.4.6 to a strong degree.
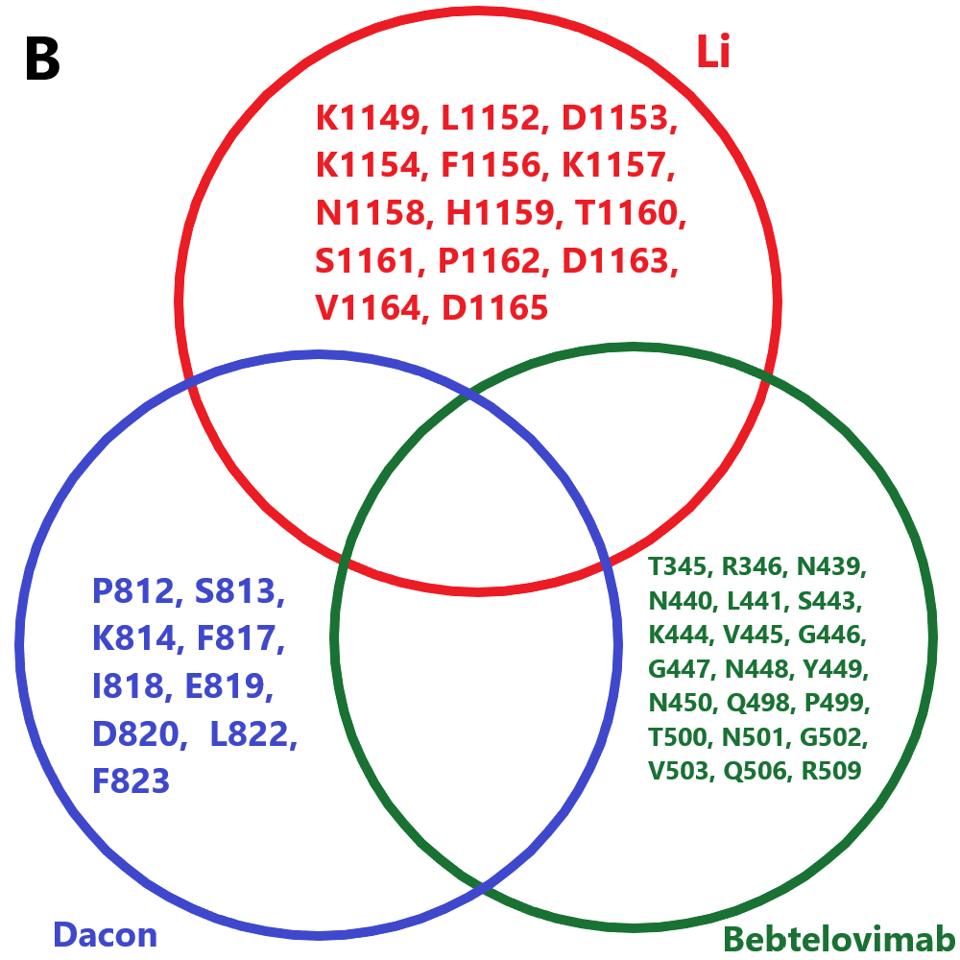
FIGURE 8: A proposed combination of antibodies for use against BA.4.6 infection.
ACCESS HEALTH INTERNATIONAL
Implications
Ultimately, we will not know the potential impact of BA.4.6 for some time. It may fade into relative inconsequence along with many variants that have come before it, but it may also emerge as the newest variant of concern. As such, we must prepare for the latter, continuing to identify and develop treatments that can neutralize the variant.

