Viroids and Virusoids: Hiding In Plain Sight
(Posted on Tuesday, July 4, 2023)
Originally published in Forbes on June 29, 2023.
This story is part of a larger series on viroids and virusoids, small infectious RNAs. You may read the others on Forbes or www.williamhaseltine.com.
Viroids and virusoids are a special kind of pathogen: small, circular strands of RNA that can infect hosts and cause disease. Think of a virus, but far more minimalist. All viruses encode at least one protein to help them transcribe and replicate their genetic material. Many others contain additional proteins that serve to evade, or even suppress, the host immune system. Barring a few exceptions, viroids and virusoids do not encode any such proteins. Despite this, they are a serious agricultural scourge, causing multi-million-dollars-worth of damage each year. Others, including hepatitis D-like agents, can infect humans and animals. New research indicates that these biological oddities may be far more common than initially thought. Published in the journal Cell, the study has led to a five-fold expansion of the known diversity of viroids and viroid-like agents.
Metatranscriptomics: Collecting Environmental RNA
Transcriptomics refers to the analysis of all the RNA content in a cell. This allows scientists to get a sense of which genes are turned on or off in cells at different points in time, including during states of infection or disease. Metatranscriptomics takes this same idea but broadens it: instead of studying the RNA content within a single cell, metatranscriptomics studies the RNA content of a much larger environmental sample. For example, marine biologists might collect a sample of seawater from a particular region to determine its RNA makeup, giving them a better sense of the microbes active in that area.
Of course, it’s not quite as simple as that. Metatranscriptomics is a complex process involving multiple steps. After initial sample collection, the RNA has to first be extracted and separated from the rest of the contents. The RNA has to then be prepared for high-throughput sequencing, which may include enrichment of desired RNA — there are various different types of RNA, some more common than others. Once the desired RNA has been sufficiently enriched, it can be sequenced. And finally, algorithms and other bioinformatic strategies are used to analyze the sequencing data.
Establishing A Pipeline
Rather than collect their own samples for analysis, Lee et al. drew upon readily-available metatranscriptome “libraries.” That is, data collected and made public by other researchers. Before they analyzed these libraries, however, they would need to establish a solid method to sift through the massive amounts of RNA and single out only those strands belonging to viroids and virusoids. Think needle in the haystack, but the haystack is the size of a football field and the lights are off.
The method—or pipeline— starts by filtering out any non-circular RNAs. This is achieved by honing in on “head-to-tail repeats”, motifs characteristic of circular RNAs and their replication intermediates. The replication intermediates are long chains of unit-length RNA tethered together, head-to-tail. These need to be cleaved from one another, so that only the unit-length RNA remains.
All of the putative circular RNAs then need to be filtered again, with a more fine-toothed comb; non-viroid circular RNAs are abundant in nature, and although we are still figuring out their exact functions, it is clear that they play an important role in gene regulation. Like most viroids and virusoids, circular RNAs do not encode any proteins. So to differentiate viroid-like circular RNAs from plain old circular RNAs, Lee et al. analyzed the sequences for the presence of self-cleaving ribozymes. They examined both the sequence itself as well as the secondary structure of the RNA molecule. Ribozymes are special sequences of RNA that, instead of encoding proteins for catalytic functions, are able to perform these functions themselves; in the case of viroids, ribozymes help cleave the long chain of tethered RNAs down to unit length, thus furthering the replication process. Think of ribozymes as “active” RNA sequences. Any potential ribozyme structures were cross-referenced against a database of known self-cleaving ribozymes
But not all viroids contain ribozymes. Members of the pospiviroidae family —one of the two main viroid families, the other being avsunviroidae— are not known to harbor ribozymes. Instead, they have a characteristic RNA section called the RY motif stem loop. It is not clear what exactly it does, but it is known to be critical to successful infection. To account for this, Lee et al. supplemented the database of known ribozymes with known RY motifs.
The last stage of the pipeline involves cross-referencing any newly discovered viroid-like RNAs against reference databases for direct comparison of sequence similarity.
Testing the Pipeline
To test their method, Lee et al. processed and searched a plant dataset composed of 1,344 transcriptomes, made up of more than 103 million full-length RNA transcripts. All viroids and viroid-like RNAs, with the exception of hepatitis D virus, infect plants. If their pipeline proved accurate, it would be able to recover known viroid-like RNAs from the transcriptomes. Using their pipeline, they predicted roughly 164,000 full-length circular transcripts. Of these, 42 were identified as viroid-like agents; 15 sequences were deemed viroid-like by virtue of having ribozymes, 33 were deemed viroid-like after cross-reference against viroid databases, and six sequences met both criteria. Their method did its job, filtering out all non viroid-RNA sequences.
The Real Thing
Next, the group of researchers applied their pipeline to 5000 diverse metatranscriptomes, totalling 1.5 billion full-length RNA transcripts. Roughly 8.5 million RNA sequences were predicted to belong to circular RNAs, with an average size of 165 nucleotides. For reference, known viroids are between 220 and 450 nucleotides long. Indeed, almost three million of the putative circular RNAs fell within the known size range of viroids. Of these, 11 thousand were classified as viroid-like because they contained a self-cleaving ribozyme. Curiously, none of the putative circular RNAs matched the RY motif of the pospiviroidae family. Ten thousand of the identified circular RNAs showed no detectable sequence similarity to known viroids, suggesting that they were newly discovered. The remaining thousand sequences were very similar to known viroids, displaying high nucleotide sequence similarity.
All in all, the metatranscriptome study yielded four thousand new sequences not listed in any viroid databases: a 5.9-fold increase in viroid-like RNA diversity.
Geographic Range
Not only are viroid-like RNAs far more diverse than initially thought, they are also a lot more widespread. Samples that came back positive for viroid-like sequences stretched the world over: Colombia, Czech Republic, Germany, USA, Japan, Malaysia, and the list goes on. Some individual sequences also spanned large geographic regions, with one sequence, for example, being found in the northern tip of Alaska, and also all the way south, in Florida.
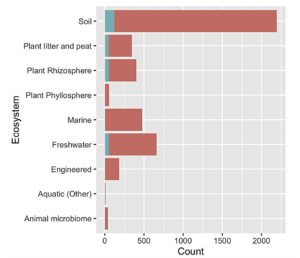
FIGURE 1. Reference (blue) and distinct clusters of viroid-like cccRNAs in different types of ecosystems. SOURCE: Lee et al. 2023 https://doi.org/10.1016/j.cell.2022.12.039
This new work by Lee et al. helps resolve a long-standing puzzle: if viroids and viroid-like RNAs are relics from a time when RNA was the dominant nucleic acid, why aren’t they more common? Well, as was made clear by this study, they are actually very common. In fact, many of the newly discovered viroid-like RNAs were among the most abundant sequences in their respective metatranscriptomes. As if a five-fold increase weren’t enough, the authors argue that this number is “likely a substantial underestimate of the true span of the viroid-like domain of the replicator space because among the millions of the predicted cccRNAs, in which no ribozymes were confidently identified, some, and possibly many, could be viroid-like agents containing unknown ribozymes or lacking ribozymes altogether, like pospiviroids.”

