Cre-LoxP: Issues Modulating Genes in the Immune System
(Posted on Saturday, January 13, 2024)

Cre-loxP, a powerful tool for preclinical cancer research, possesses intrinsic pitfalls that can alter result accuracy, according to an editorial paper in Oncoscience.
GETTY
Cancer and other diseases could be treated by turning certain genes on and off in the immune system. The method used to activate and inactivate these genes is critical to consider. If the technique does not pinpoint the right cells or lingers in the body for too long, the patient can incur damage. For example, disabling regulatory T cell activity in cancer models can encourage antitumor activity; however, if sustained for too long, the body can lose the ability to recognize its own cells and autoimmune reactions can result.
Cre-loxP, a system commonly wielded to manipulate gene expression in mammals, may not be the right fit. In their editorial paper published in Oncoscience, researchers Piotr Czarnota and Jaroslaw Cisowsk highlight several potential limitations of this powerful system called Cre-loxP. Given their careful warnings, it may be more valuable to entertain other gene regulation methods in preclinical experiments.
How Cre-loxP Works
Cre-loxP is a technology used to manipulate gene expression in animal cells through selective breeding, particularly in mice. It relies on two components: an enzyme called Cre recombinase and specific DNA recognition sites called loxP. Researchers manipulate loxP sites to flank a gene of interest. When the enzyme recognizes the loxP sites and cuts at these locations, the gene of interest is either excised, inverted, deleted or translocated in the process.
Gene expression can be edited with further precision using promoters. Promoters are DNA sequences located upstream of a gene that play a crucial role in initiating the transcription of a particular gene. In Cre-loxP, they can determine where and when Cre recombinase is transcribed and activated. A cell-specific promoter can restrict expression to a tissue-specific location, while an inducible promotor allows the enzyme to activate only after a certain trigger is introduced.
The power of this system is exemplified through a paper published in PNAS. The work by Bert O’Malley and colleagues details how altering the gene expression of a protein in regulatory T cells can significantly impact the fate of mice with breast and prostate tumors. Cre-loxP is used to breed mice that do not possess steroid receptor coactivator 3 (SRC-3, pronounced Sark Three), a protein that is highly involved in regulatory T cell activity. The SRC-3 gene knockout is observed only after a drug called tamoxifen is injected into the mice, a step that temporally induces Cre enzyme activity.
Normal mice and the knockout mice then receive an injection of breast cancer cells. When analyzed 20 days later, the normal mice experienced rapid tumor growth; the knockout mice, in comparison, demonstrated undetectable tumor growth for more than 200+ days. This result is mirrored when repeated with prostate cancer cells. The study demonstrates how interrupting regulatory T cell suppression through SRC-3 knockout can allow other immune cells to fight tumors effectively.
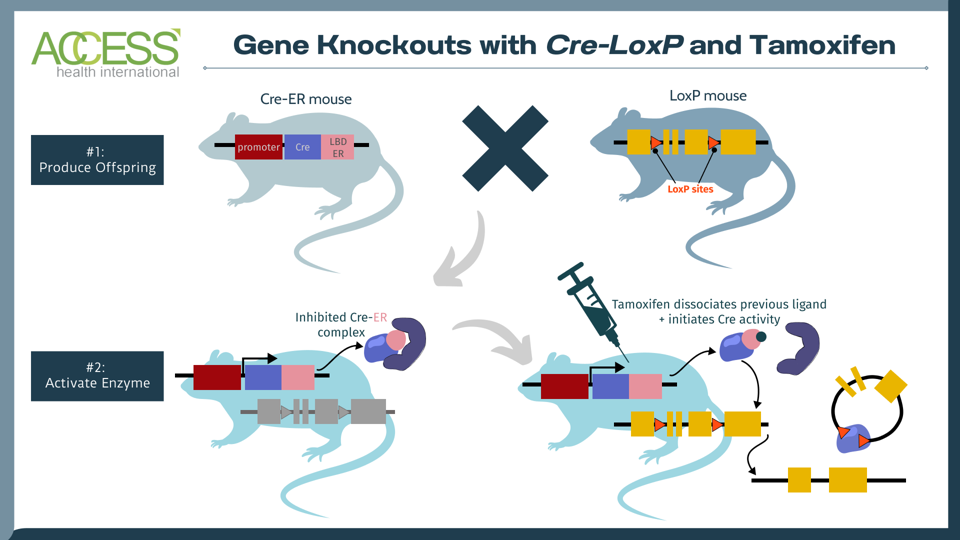
FIGURE 1: The tamoxifen-induced Cre-loxP system is a technique used to knock out a gene of interest in mammals. One set of mice contains the tissue-specific instructions to produce Cre, an enzyme that recognizes a specific DNA sequence called LoxP. Another set of mice is edited to express a gene of interest flanked by LoxP sites. When these two sets of mice reproduce, their offspring contain the genetic instructions for Cre, LoxP and the gene of interest. The Cre recombinase enzyme activates once the offspring receives a tamoxifen injection. The enzyme then recognizes the LoxP DNA sites in the offspring, cuts at these two locations and knocks out the gene of interest. The result: offspring mice without the gene of interest. Abbreviations: LDB ER, estrogen receptor with mutated ligand binding domain.
ACCESS HEALTH INTERNATIONAL
Potential Limitations
Although Cre-loxP is an undoubtedly valuable tool for observing gene changes in living animals, the system also carries certain limitations that may hinder the accuracy of the results. In their editorial paper, authors Piotr Czarnota and Jaroslaw Cisowsk emphasize certain challenges that may be overlooked when using Cre-loxP for cancer research. Carefully considering these warnings may lead researchers to more meticulous Cre-loxP experimental designs and interpretations.
The first highlighted limitation revolves around the fidelity of promoters used in Cre-loxP. While certain promoters can spatially or temporally restrict Cre expression to select tissues, this specificity relies on the cell type fidelity of the selected promoter. Cell type fidelity refers to the promoter’s accuracy and reliability to drive Cre expression in desired cell types. The authors warn that if the promotor is not specific enough to a certain cell type and drives Cre expression in other cell types as well, this can lead to inaccurate or unintended genetic changes. For example, many promoters specific to cells in the pancreas are also expressed in other cells at early stages of development, including some brain neurons and cells in the liver, stomach and intestines. The genetic recombination observed as a result may lack specificity.
Confusion could also arise when microvesicles transfer Cre mRNA into neighboring cells. These nearby cells do not actually possess the desired characteristics or gene expression patterns and yet receive the Cre mRNA. As the target population of cells is not highlighted, this can lead to a false interpretation of results.
Additionally, researchers must consider the more plastic nature of cancer cells when using Cre-loxP. Cancer cells can acquire characteristics from other cell lineages due to the expression of oncogenes or deletion of tumor suppressor genes. This means that it is possible for promoters to be abnormally activated in cells they are not normally active in. This may distort where Cre expression appears to originate from.
Another challenge that threatens the proper interpretation of results is a temporary induction of Cre recombinase expression in unintended cell types. This process can occur due to various processes, but ultimately, the loxP sites may be unintentionally deleted in these cells, and these erroneous changes can passed onto offspring/future generations of cells. In response, researchers may mistakenly conclude that the genetic change occurred in the intended cell; in reality, unintended genetic changes occurred in unintended cell types. The authors note two processes that can yield this erroneous result: transdifferentiation, when the gene expression pattern characteristic of one cell type is changed for that of another cell type; and epithelial-mesenchymal transition (EMT), when epithelial cells lose key characteristics and gain more mesenchymal traits, including enhanced migratory properties.
Research Implications
Cre-loxP provides a simple yet potent avenue for researchers to visualize modified gene expression in living animal models. Just this year, this platform proved paramount in discovering how SRC-3 knockout can lift regulatory T cell suppression and encourage tumor-fighting activity in the immune system. However, the system possesses intrinsic pitfalls, such as the potential loss of fidelity of Cre recombinase expression and the risk of erroneous conclusions due to unintended genetic changes. It may be better to entertain other technologies such as CRISPR/Cas9 gene editing to alter gene expression in experiments, as these methods are proven to be precise, practical and largely safe when applied to humans.
This article was originally published on Forbes and can be read online here.

