Attack and Counterattack: How SARS-CoV-2 Blocks Our Natural Immune Defenses
(Posted on Monday, June 20, 2022)
This article is an extension of our series on immune suppression by SARS-CoV-2. The series has since been published as a book, Natural Immunity and Covid-19: What it is and How it Can Save Your Life. It is also available to read on my website. Here, we discuss new data regarding the ORF7a viral protein.
During the initial stages of cellular infection, SARS-CoV-2 releases a number of accessory proteins to help suppress and evade our immune system. ORF7a is one such protein. Suppression of the innate immune system is necessary for the virus to establish infection. The several-day suppression of the immune system by the virus also contributes to the asymptomatic incubation period. During this period the virus can be transmitted from person-to-person before anyone is even aware they have been infected. ORF7a is a key player in preventing the phosphorylation of a host protein called signal transducer and activator of transcription 2 (STAT2), which is critical for the induction of antiviral interferon-stimulated genes. Novel research highlights additional means by which ORF7a contributes to the infectivity, pathogenesis, and transmission of SARS-CoV-2. Published in Nature Communications, the work by Timilsina et al. details how ORF7a counteracts the protective effects of serine incorporator 5 (SERINC5), a host protein that blocks viral entry into cells.
On the Offensive: SERINC5
The antiviral functions of SERINC5 against retroviruses, including human immunodeficiency virus 1 (HIV-1) and murine leukemia virus (MLV), are well-documented. Yet little is known about its role in coronavirus infections. Timilsina and his colleagues set out to fill this gap in our knowledge.
They first examined the expression levels of each member of the SERINC gene family —one through five— in lung tissue and Calu-3 lung cells. All except SERINC4 were abundantly expressed. Infection with SARS-CoV-2 did not influence the expression of SERINC1,2,3, and 5 in the lung tissue or the Calu-3 lung cells.
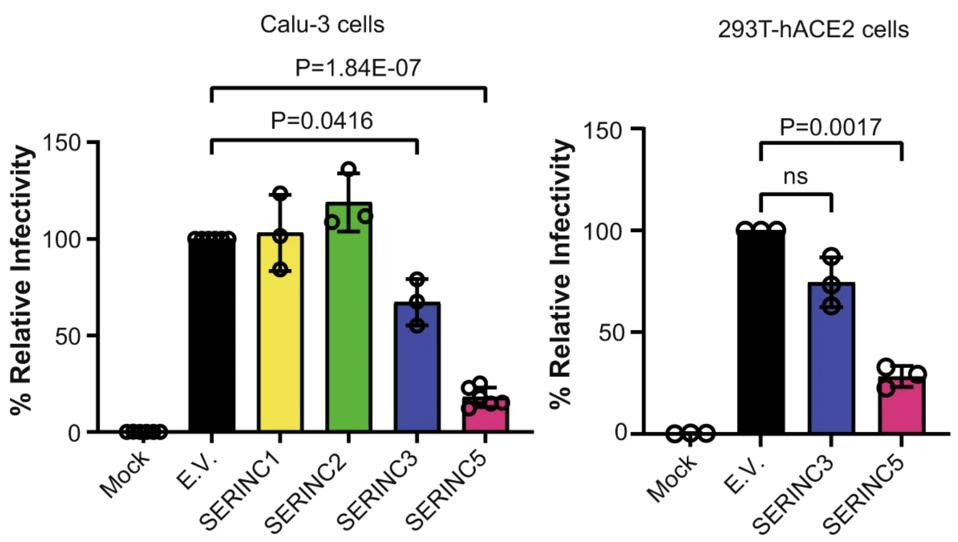
Next, the researchers examined if any of the SERINC genes expressed in the lung tissue exhibit protective effects similar to those seen with SERINC5 in HIV-1. To do so, they produced pseudoviruses that replaced the HIV-1 exterior protein with the SARS-CoV-2 Spike protein —which the virus uses to bind to host receptors and enter into cells. All SERINC proteins were absorbed into the SARS-CoV-2 Spike pseudovirions. When exposed to these SERINC-containing pseudovirions, lung cells and kidney cells were significantly less likely to become infected. SERINC5 proved to be especially effective at reducing infectivity in both cell types (Figure 1). SERINC3 modestly reduced viral infectivity. SERINC1 and SERINC2 had no effect.
FIGURE 1. Relative infectivity of lung cells (Calu-3) and kidney cells (293T-hACE2) when exposed to … [+] FROM: TIMILSINA ET AL. 2022
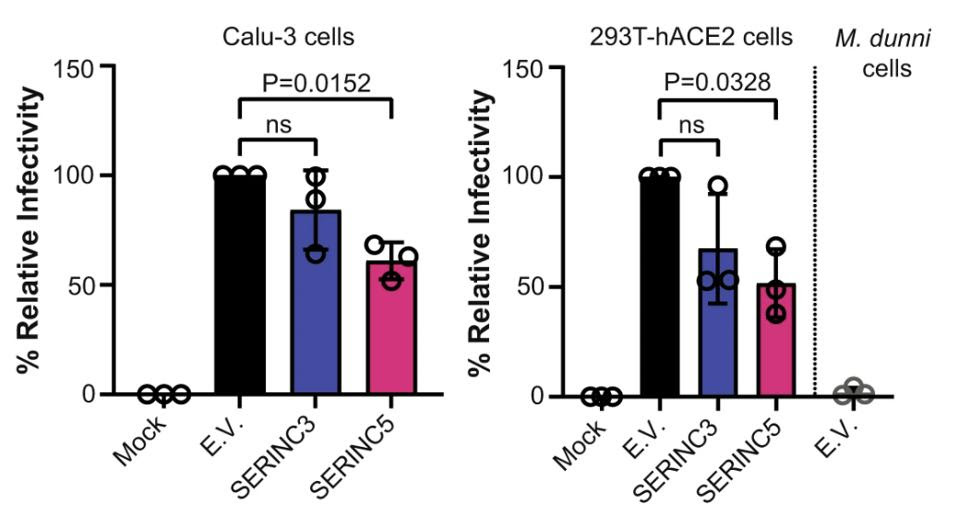
But retroviruses and coronaviruses are quite different from one another. For one, they assemble in different parts of the host cell; retroviruses in the plasma membrane and coronaviruses in the endoplasmic-reticulum-Golgi intermediate compartment (ERGIC). The pseudoviruses the researchers generated may not accurately represent what happens with coronaviruses. To remedy this, they performed the same experiments on both SARS-CoV-2 virus-like particles (VLPs) —non-infectious replicas of the virus— and infectious SARS-CoV-2. As before, SERINC5 successfully incorporated itself into the Spike protein and successfully reduced viral infectivity (Figure 2). SERINC3 was incorporated into the infectious SARS-CoV-2, but had a much smaller impact on viral infectivity.
FIGURE 2. Relative infectivity of lung cells (Calu-3) and kidney cells (293T-hACE2) when exposed to … [+] FROM: TIMILSINA ET AL. 2022
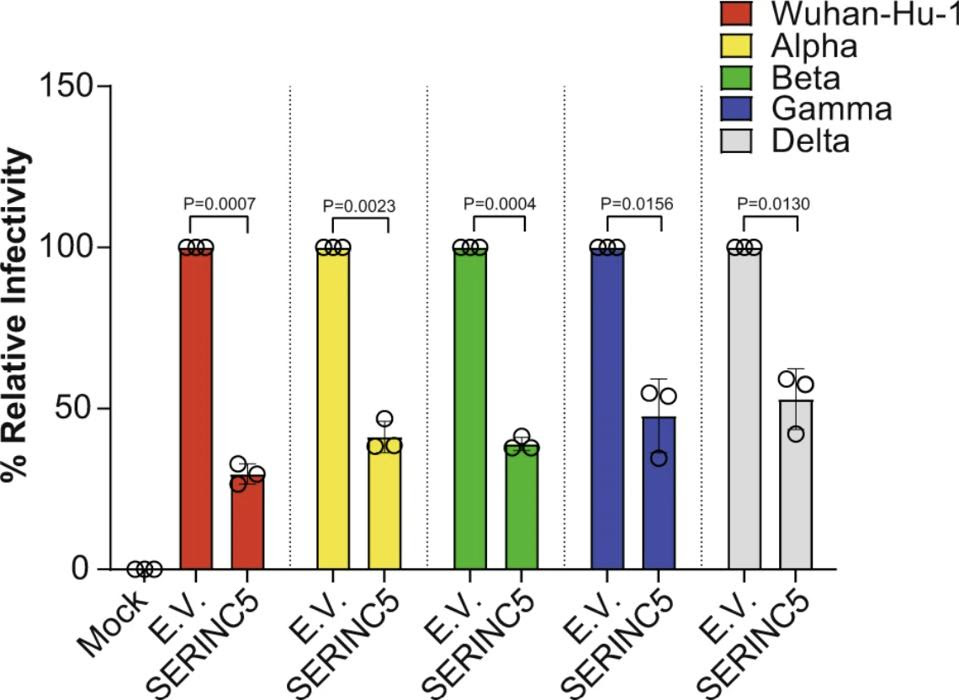
The researchers also tested for SERINC5 incorporation into the Spike protein of different SARS-CoV-2 variants —Alpha (B.1.1.7), Beta (B.1.351), Gamma (P1), and Delta (B.1.617)— to make sure any mutations to the Spike protein did not negate the protective effects to SERINC5. Across all variants, SERINC5 continued to restrict viral infectivity (Figure 3).
FIGURE 3. Relative infectivity of kidney cells (293T-hACE2) when exposed to different variants of … [+] FROM: TIMILSINA ET AL. 2022
So SERINC5 gets incorporated into the Spike protein of SARS-CoV-2 and, from there, manages to reduce infection. But, how exactly does it accomplish this?
Timilsina et al. initially suspected that SERINC5 interferes with receptor binding, the first step of the process through which SARS-CoV-2 gains entry into our cells. This happens when SARS-CoV-2 uses its Spike protein to bind to angiotensin converting enzyme 2 (ACE2) receptors on the outside of our cells. Blocking this interaction would block the possibility of infection. To the surprise of the researchers, SERINC5 had no impact on the interaction between the Spike protein and our cells’ ACE2 receptors.
Nor did SERINC5 interfere with the next stage of cellular infection — cleavage of the Spike protein into two parts.
Once the SARS-CoV-2 Spike protein has bound to ACE2 and has been cleaved, the remaining section of the Spike protein inserts itself into the host cell membrane and pulls itself in, fusing the two together. Timilsina et al. discovered that, in the presence of SERINC5, the Spike-mediated fusion to the host cell membrane was noticeably reduced. This suggests SERINC5 blocks viral entry by interfering with the fusion stage of infection. The precise mechanism by which it does this remains to be determined.
Viral Counterattack: ORF7a
HIV-1 has developed a way of parrying the blow dealt by SERINC5; it encodes a protein called Nef that prevents the antiviral protein from being incorporated into the budding virions. What about SARS-CoV-2, does it wield any counterattacks of its own?
The group of researchers turned to SARS-CoV-2 accessory proteins, known to block host antiviral genes and suppress the immune response. They honed in on ORF7a. During infection with SARS-CoV-2, ORF7a moves to the endoplasmic reticulum and Golgi apparatus of the host cell. This is the area where new viral particles are assembled and, by extension, where SERINC5 may incorporate itself into the nascent Spike protein.
Timilsina et al. tested their hypothesis by exposing lung and kidney cells to a knock-out strain of SARS-CoV-2 that does not contain ORF7a. They compared the results to infection with unmutated wild-type SARS-CoV-2. The amount of SERINC5 packaged into the budding virions was significantly increased in the ORF7a knock-out strain. Higher levels of SERINC5 in the knock-out strain were reflected by poorer viral infectivity overall. Reintroducing ORF7a to the knock-out strain salvaged infectivity. These findings confirm that, in the context of SARS-CoV-2 infection, ORF7a works to prevent the incorporation of SERINC5 into nascent viral particles.
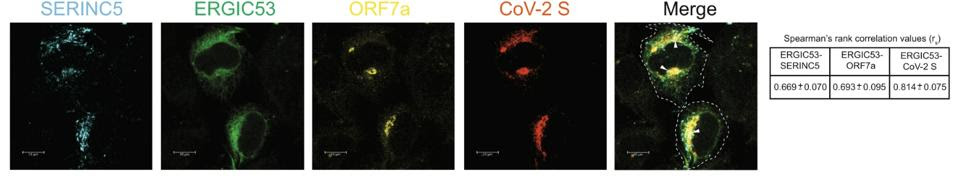
Timilsina and his colleagues propose two methods through which ORF7a inhibits SERINC5. One method is by preventing SERINC5 from being packaged into the nascent virion particles in the first place. The second method happens within the viral particles. The researchers suggest ORF7a forms a complex with the Spike protein and with SERINC5 that ultimately blocks SERINC5 from restricting viral entry. They were able to confirm that the SARS-CoV-2 Spike protein, ORF7a, and SERINC5 all come together and interact at the endoplasmic-reticulum–Golgi intermediate compartment (ERGIC) (Figure 4). The nuances of how this complex undermines SERINC5 remain unknown. By analogy to HIV-1, the authors hypothesize that SERINC5 may alter the structure of the Spike protein and that ORF7a binds to the Spike protein to prevent any such changes. Future research should aim to resolve this unknown through in-depth structural analysis.
FIGURE 4. Co-location of SERINC5, ORF7a, and SARS-CoV-2 Spike protein at the ERGIC. FROM: TIMILSINA ET AL. 2022
Mutations to accessory proteins are not rare and a number of naturally occurring deletions have been detected in ORF7a; do any of them undermine its ability to mount a counterattack against SERINC5?
Timilsina et al. tested four naturally occurring deletions of ORF7a — Δ9nt, Δ18nt, Δ57nt, and Δ96nt— isolated from clinical samples of infected patients. The ability to block SERINC5 was preserved across all four of the SARS-CoV-2 variants with naturally occurring ORF7a mutations. There was little to no difference in the extent of SERINC5 restriction between the ORF7a mutations and the wild-type ORF7a.
Implications
Of more general importance is that inhibition of ORF7a will weaken SARS-CoV-2 replication, allowing our natural cellular defenses to be more effective in warding off the virus. This work by Timilsina et al. adds another reason to consider ORF7a an important antiviral target for future drug development.