Solving The Question Of Covid Variant Increased Fitness Is Like Deciphering A Rubik’s Cube
(Posted on Thursday, August 25, 2022)
A hallmark of the Covid-19 pandemic is the successive waves of infection with one variant after another. Each variant appears more fit than its predecessor in terms of its ability to infect and spread in a population.
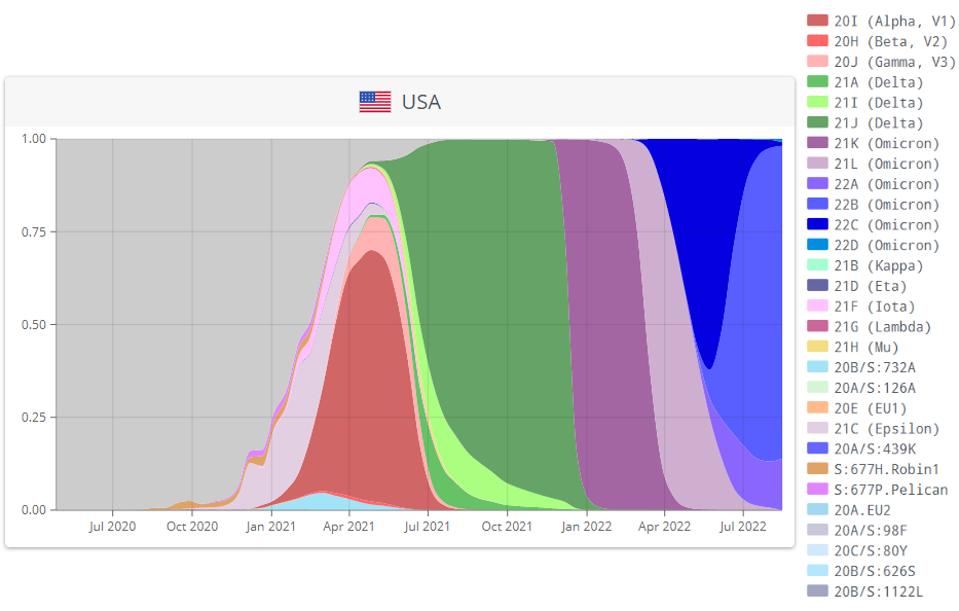
FIGURE 1: Successive waves of SARS-CoV-2 variants in the Covid-19 pandemic.
COVARIANTS.ORG
FIGURE 1: Successive waves of SARS-CoV-2 variants in the Covid-19 pandemic.
The ability of one virus to replace another is determined by its ability to evade the immune system of those previously infected or vaccinated, as well as intrinsic replication properties. It is essential to distinguish between these two properties as they work cooperatively to determine how fast a variant spreads through a population.
Most studies throughout the pandemic have focused on the ability of the virus to evade the immune system because of the importance of prior immunity. However, a few studies have investigated the intrinsic replication properties of the virus. Two recent studies have revealed previously unknown epidemiological properties.
The first by Wu et al. revealed that the incubation periods of successive variants are growing shorter. Their meta-analysis of 141 studies throughout the pandemic showed that the average incubation period of all cases was 6.57 days. For the Alpha variant specifically, the period was 5.00 days. For Beta, it was 4.50 days. For Delta, it was 4.41 days. For Omicron, it was only 3.42 days. This means later variants get the host sick sooner after the initial infection, and the virus moves much quicker from host to host.
A second paper by Reuschl et al. demonstrated that the ability of the virus to suppress interferon, the first line of defense for innate immunity, is increased amongst the latest variants.
These two properties are most likely associated with proteins other than the Spike. Specifically, the non-Spike structural proteins, namely E, M, and N, and the accessory proteins Orf3a through Orf10.
In a series of elegant experiments described by Syed et al., the Gladstone Institutes have examined the intrinsic replicative properties of the virus in the absence of the Orf proteins using a simplified virus-like particle. Virus-like particles are constructed only to contain specific proteins, allowing isolated testing of viral characteristics such as replication using luciferase enzymes to measure results.
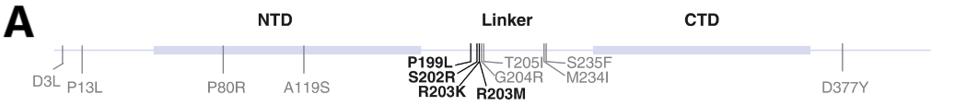
In a previous study, the same group of researchers found that the N protein was critical to the virus replication of the Alpha variant. They found that the four mutations in most natural variants increase mRNA delivery and expression by about 10-fold, whereas other mutations such as R203M increase virus production between 50 to 150-fold, displaying the significant role the structural proteins can play in transmission pathogenesis.
SYED ET AL.
FIGURE 2: N Protein Enhanced Replication. (A) Map of SARS-CoV-2 N domains showing the locations of observed mutations. Mutations that were observed to enhance signal are shown in bold. (B) RT-qPCR of supernatant collected from A549-ACE2 cells infecte
SYED ET AL.
In their recent study, they expand to analyze more recent variants and look at the effects of other proteins on replication outcomes, specifically, the structural proteins E, M, N, and S.
Using virus-like particles, they could individually examine the impact of mutations in each structural protein. First investigating the S protein, they found that the infectivity of the Delta Spike drops significantly compared to the ancestral B.1, whereas the Omicron Spike is roughly equivalent.
Next, they examined the N protein of the three viruses and B.1.1, which includes mutations R203K and G204R. The B.1.1 N is roughly six-fold more infectious than B.1, Delta N is 23.5-fold more infectious, and Omicron N is 26-fold more infectious, indicating the heavily mutated Delta and Omicron N proteins contribute significantly to virus infectivity.
Syed et al. then tested the M and E proteins, which is conjunction drop infectivity of both Delta and Omicron four-fold as compared to B.1. However, the Omicron M protein on its own remained as infectious as the B.1 isolate, indicating that the Omicron E protein contributes dropping infectivity.
Finally, they analyzed the different viruses with a complete set of S, N, E, and M proteins. B.1.1 was 5.3-fold more infectious than the ancestral B.1 strain, whereas Delta was only 3.7-fold more infectious, and Omicron was 4.6-fold more infectious (Figure 2).
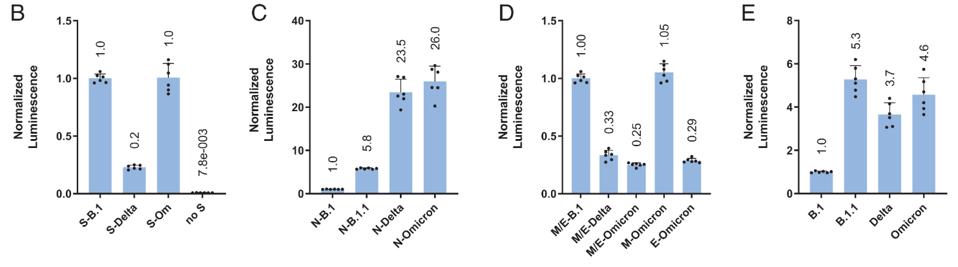
FIGURE 3: Omicron structural gene variants alter the infectivity of SC2-VLPs. SC2-VLPs assembled in packaging cells transformed with plasmids encoding S, E, M, and N genes and a luciferase mRNA fused to the SARS-CoV-2 packaging signal are tested for
SYED ET AL.
They also found that the cleavage and processing of Spike varied both between different S variants and when coexpressed with differing M and N variants. For example, Spike was less efficiently cleaved when all S, N, M, and E, were Omicron variants rather than an isolated Omicron Spike (Figure 3).
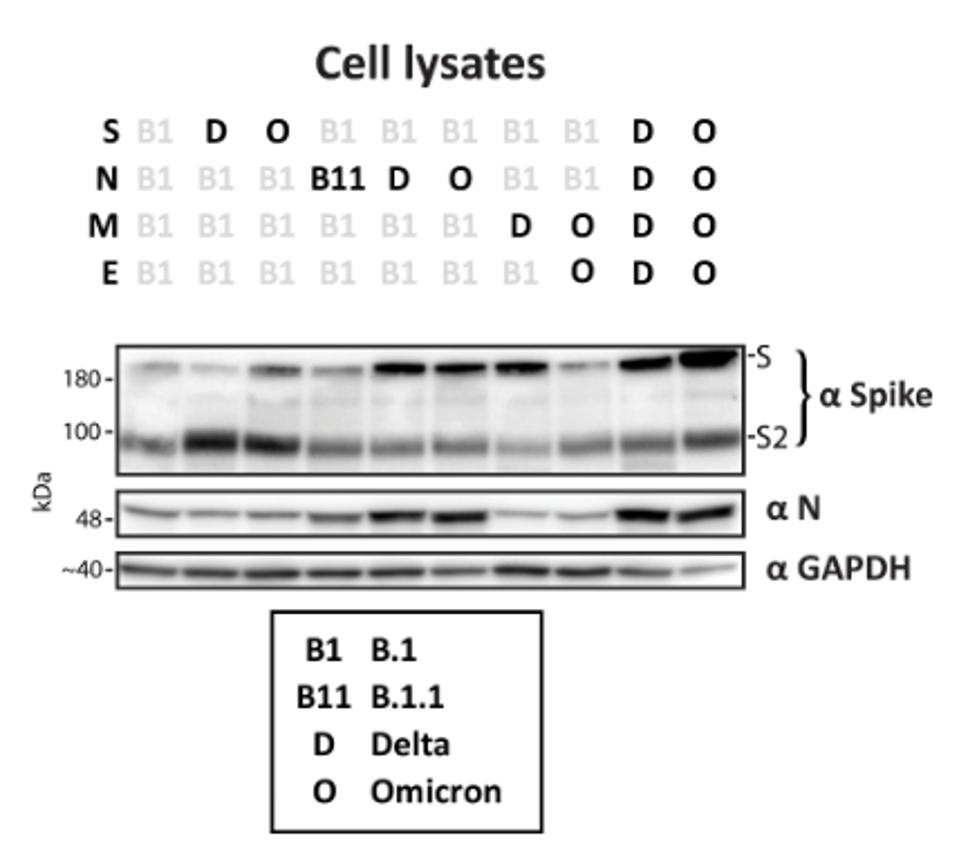
FIGURE 4: Western blot of cell lysates from cells transfected to generate VLPs stained for N, S, and GAPDH as a loading control.
SYED ET AL.
These results amplify the notion that N protein mutations play an essential role in the increased fitness of the virus. They also reveal complex interactions between the structural proteins, suggesting epistatic effects of mutated viral proteins working together to create a more infectious virus. For instance, introducing E, M, and N to an S protein may increase overall viral expression, or in some cases, it may even reduce the replication level.
These experiments add substantially to our understanding of the fitness of variants and how one virus can displace another. It involves not only immune evasion but also the ability of the virus to replicate and compensatory mutations throughout the genome that contribute to the virus’s ability to spread throughout a population.
To solve a Rubik’s Cube, you must have the nine pieces of each of the six sides correctly aligned. This is a good analogy for viral fitness. In addition to having optimal mutations in the Spike protein, there must also be optimal mutations in the E, M, and N proteins as well. In addition, there are many more dimensions to consider, as SARS-CoV-2 has roughly 30 proteins throughout the genome, akin to an immensely complicated 30-sided Rubik’s Cube.
We await further studies that include the effects of accessory genes that are well known to play a role in the ability of the virus to counteract the immune system and possibly increase the rate of virus spread. This information would be critical to understanding current variants and what to expect from future iterations that may be more fit and possibly more dangerous or virulent.

